NPs Basic Information
|
Name |
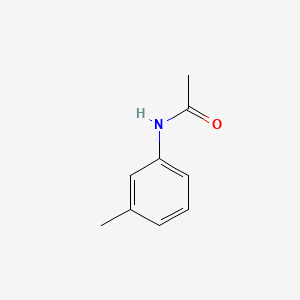
3'-Methylacetanilide
|
| Molecular Formula | C9H11NO | |
| IUPAC Name* |
N-(3-methylphenyl)acetamide
|
|
| SMILES |
CC1=CC(=CC=C1)NC(=O)C
|
|
| InChI |
InChI=1S/C9H11NO/c1-7-4-3-5-9(6-7)10-8(2)11/h3-6H,1-2H3,(H,10,11)
|
|
| InChIKey |
ALMHSXDYCFOZQD-UHFFFAOYSA-N
|
|
| Synonyms |
3'-Methylacetanilide; 537-92-8; N-Acetyl-m-toluidine; 3-Methylacetanilide; m-Acetotoluide; m-Acetotoluidide; N-(3-METHYLPHENYL)ACETAMIDE; m-Methylacetanilide; m-Acetotoluidine; m-Tolylacetamide; 3-Acetamidotoluene; N-m-Tolylacetamide; Aceto-m-aminotoluene; Acetamide, N-(3-methylphenyl)-; Acetotoluide; m-Acetotolidide; N-Acetyl-3-methylaniline; 1-Acetamido-3-methylbenzene; 3-ACETYLAMINOTOLUENE; NSC 3103; KY86R0888B; NSC-3103; m-Methyl acetanilide; N-Acetyl-n-toluidine; N-Acetoxy-3-toluidine; CCRIS 5955; EINECS 208-678-1; N1-(3-methylphenyl)acetamide; UNII-KY86R0888B; AI3-16907; meta-Acetotoluidide; 3\'-Methylacetanilide; DSSTox_CID_4412; DSSTox_RID_77391; M-ACETOTOLUIDE [MI]; DSSTox_GSID_24412; SCHEMBL12133; WLN: 1VMR C1; 3'-Methylacetanilide, 98%; MLS002415752; N-(3-Tolyl)acetic acid amide; N-ACETYL-META-TOLUIDINE; ACETYL-M-TOLUIDINE, N-; CHEMBL1528164; DTXSID6024412; NSC3103; HMS3039L06; ZINC153024; Tox21_200887; MFCD00014962; STK301244; AKOS003870217; AB01179; NCGC00091308-01; NCGC00091308-02; NCGC00258441-01; CAS-537-92-8; LS-13628; SMR001370913; DB-013357; A0062; CS-0204499; FT-0616116; D88205; W-105708; Q27282500; Ethyl3-(trifluoromethyl)-1,2,4-oxadiazole-5-carboxylate
|
|
| CAS | 537-92-8 | |
| PubChem CID | 10843 | |
| ChEMBL ID | CHEMBL1528164 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 149.19 | ALogp: | 1.7 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 29.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.653 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.32 | MDCK Permeability: | 0.00002970 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.288 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.847 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.92 | Plasma Protein Binding (PPB): | 38.90% |
| Volume Distribution (VD): | 1.082 | Fu: | 54.68% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.933 | CYP1A2-substrate: | 0.934 |
| CYP2C19-inhibitor: | 0.49 | CYP2C19-substrate: | 0.839 |
| CYP2C9-inhibitor: | 0.07 | CYP2C9-substrate: | 0.452 |
| CYP2D6-inhibitor: | 0.045 | CYP2D6-substrate: | 0.742 |
| CYP3A4-inhibitor: | 0.042 | CYP3A4-substrate: | 0.528 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.128 | Half-life (T1/2): | 0.781 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.033 | Human Hepatotoxicity (H-HT): | 0.539 |
| Drug-inuced Liver Injury (DILI): | 0.53 | AMES Toxicity: | 0.194 |
| Rat Oral Acute Toxicity: | 0.029 | Maximum Recommended Daily Dose: | 0.07 |
| Skin Sensitization: | 0.672 | Carcinogencity: | 0.164 |
| Eye Corrosion: | 0.053 | Eye Irritation: | 0.852 |
| Respiratory Toxicity: | 0.031 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000239 | 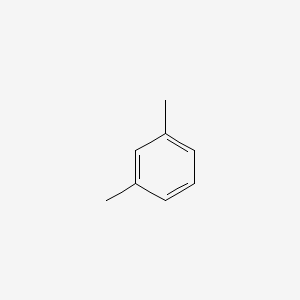 |
0.486 | D0U5QK | 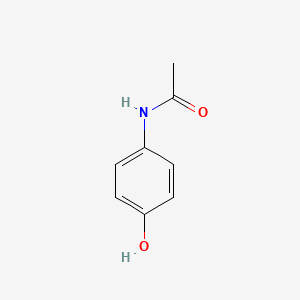 |
0.463 | ||
| ENC000072 | 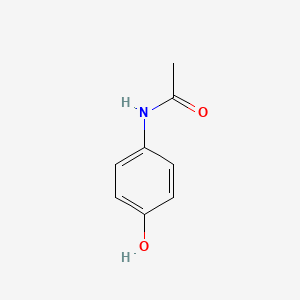 |
0.463 | D02AQY | 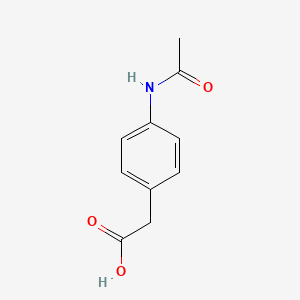 |
0.388 | ||
| ENC000414 | 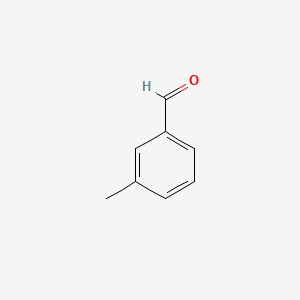 |
0.447 | D01PJR | 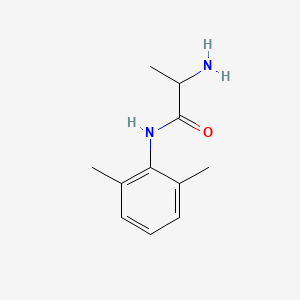 |
0.340 | ||
| ENC000413 | 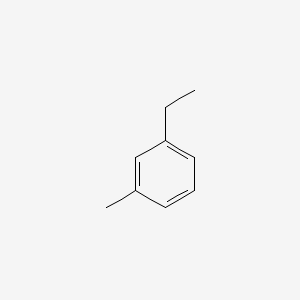 |
0.447 | D0J9XZ | 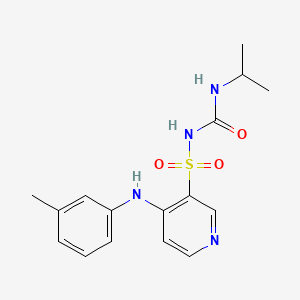 |
0.333 | ||
| ENC000106 | 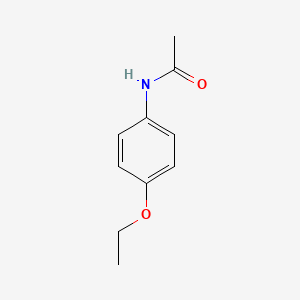 |
0.435 | D0KD1U | 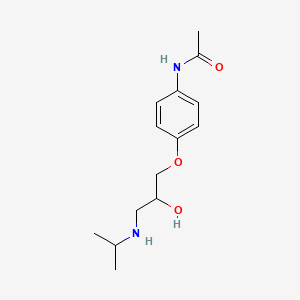 |
0.323 | ||
| ENC000368 | 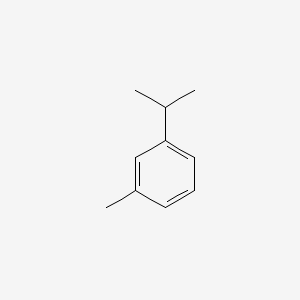 |
0.425 | D06MRT | 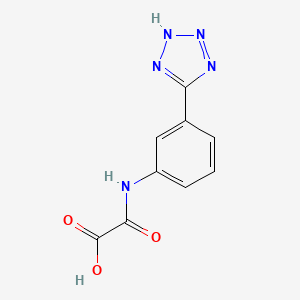 |
0.322 | ||
| ENC002213 | 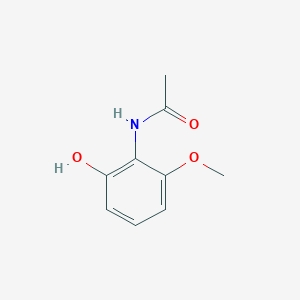 |
0.413 | D0X4ZR | 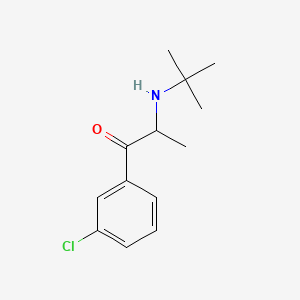 |
0.309 | ||
| ENC000391 | 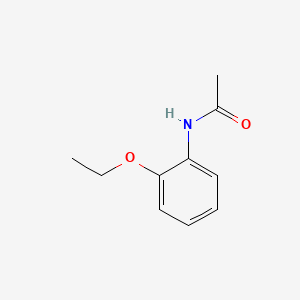 |
0.404 | D06LYG | 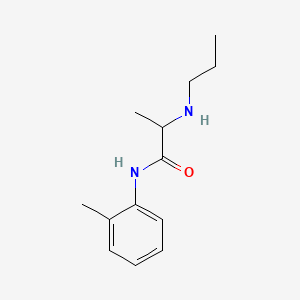 |
0.298 | ||
| ENC000612 | 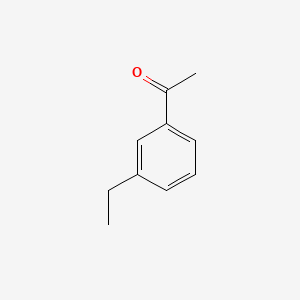 |
0.395 | D0M4VM | 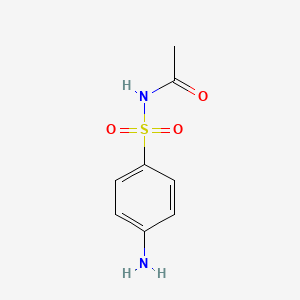 |
0.288 | ||
| ENC002891 | 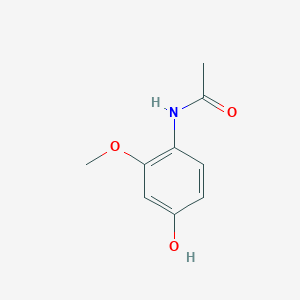 |
0.383 | D0X4RN | 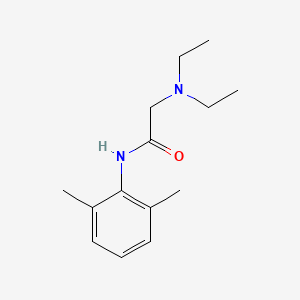 |
0.288 | ||