NPs Basic Information
|
Name |
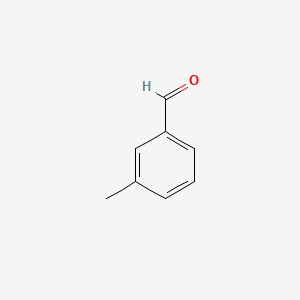
3-Methylbenzaldehyde
|
| Molecular Formula | C8H8O | |
| IUPAC Name* |
3-methylbenzaldehyde
|
|
| SMILES |
CC1=CC(=CC=C1)C=O
|
|
| InChI |
InChI=1S/C8H8O/c1-7-3-2-4-8(5-7)6-9/h2-6H,1H3
|
|
| InChIKey |
OVWYEQOVUDKZNU-UHFFFAOYSA-N
|
|
| Synonyms |
3-Methylbenzaldehyde; M-TOLUALDEHYDE; 620-23-5; Benzaldehyde, 3-methyl-; 3-tolualdehyde; m-Methylbenzaldehyde; 3-Tolylaldehyde; m-Tolyl aldehyde; m-Toluylaldehyde; 3-methyl benzaldehyde; 3-methyl-benzaldehyde; M-Tolualdehyde, stabilized; MFCD00003374; OWH6650C4Y; NSC-1244; NSC-89859; 3-Methyl-Benzaldehyd; NSC 1244; EINECS 210-632-0; NSC 89859; UNII-OWH6650C4Y; m-tolylaldehyde; metatolualdehyde; AI3-02278; HSDB 7691; m-toluic aldehyde; m-Tolualdehyde, b; meta-tolyl aldehyde; m-methyl benzaldehyde; meta-methylbenzaldehyde; M-FORMYLTOLUENE; m-Tolualdehyde, 97%; TOLUALDEHYDE,M-; META TOLUALDEHYDE; TOLUALDEHYDE, M-; bmse000552; NCIOpen2_001577; 3-CH3C6H4CHO; SCHEMBL65797; 3-Methylbenzaldehyde, stab. with 0.1% hydroquinone; CHEMBL4475423; DTXSID6060717; BDBM85649; CHEBI:28476; NSC1244; ZINC896722; 3-METHYLPHENYLCARBOXALDEHYDE; m-Tolualdehyde, analytical standard; 3-METHYLBENZALDEHYDE [HSDB]; NSC89859; FEMA NO. 3068, M-; AC8308; STL194066; AKOS000119450; AC-2438; AS-14544; HY-78086; SY001146; CS-0007820; FT-0629011; T0258; EN300-20440; C07209; A833543; J-512886; Q26828654; F2190-0581; Z104478208; 3-methylbenzaldehyde;3-Methylbenzaldehyde, stab. with 0.1% hydroquinone
|
|
| CAS | 620-23-5 | |
| PubChem CID | 12105 | |
| ChEMBL ID | CHEMBL4475423 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 120.15 | ALogp: | 2.0 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.52 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.316 | MDCK Permeability: | 0.00002610 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.018 |
| Human Intestinal Absorption (HIA): | 0.007 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.987 | Plasma Protein Binding (PPB): | 81.58% |
| Volume Distribution (VD): | 0.999 | Fu: | 16.95% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.959 | CYP1A2-substrate: | 0.636 |
| CYP2C19-inhibitor: | 0.426 | CYP2C19-substrate: | 0.57 |
| CYP2C9-inhibitor: | 0.082 | CYP2C9-substrate: | 0.586 |
| CYP2D6-inhibitor: | 0.012 | CYP2D6-substrate: | 0.624 |
| CYP3A4-inhibitor: | 0.026 | CYP3A4-substrate: | 0.255 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.903 | Half-life (T1/2): | 0.727 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.052 | Human Hepatotoxicity (H-HT): | 0.028 |
| Drug-inuced Liver Injury (DILI): | 0.039 | AMES Toxicity: | 0.034 |
| Rat Oral Acute Toxicity: | 0.014 | Maximum Recommended Daily Dose: | 0.052 |
| Skin Sensitization: | 0.287 | Carcinogencity: | 0.047 |
| Eye Corrosion: | 0.989 | Eye Irritation: | 0.996 |
| Respiratory Toxicity: | 0.973 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000239 |  |
0.516 | D0E9CD |  |
0.410 | ||
| ENC000413 |  |
0.471 | D06GIP | 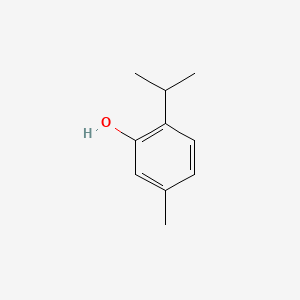 |
0.286 | ||
| ENC000012 | 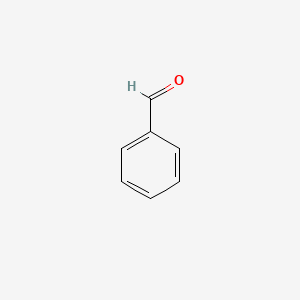 |
0.455 | D01ZJK | 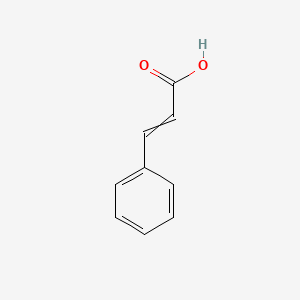 |
0.273 | ||
| ENC000370 | 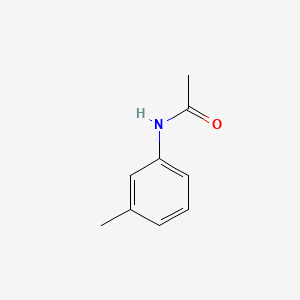 |
0.447 | D0S5LH | 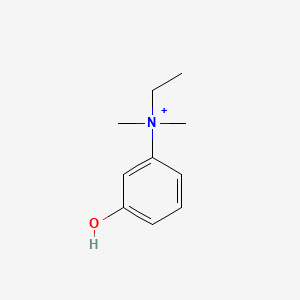 |
0.267 | ||
| ENC000649 | 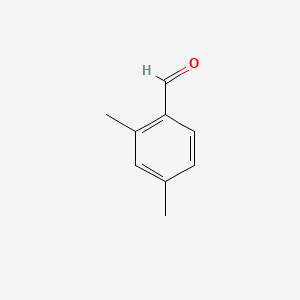 |
0.444 | D04EYC | 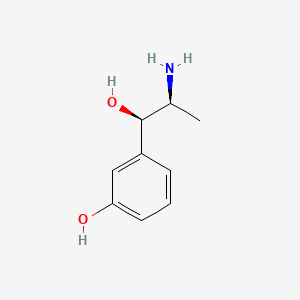 |
0.267 | ||
| ENC000552 | 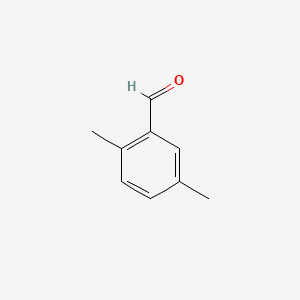 |
0.444 | D0O6IU | 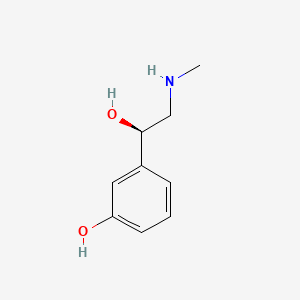 |
0.261 | ||
| ENC000368 | 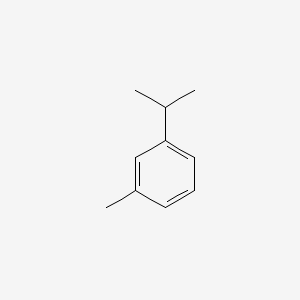 |
0.444 | D04YMH | 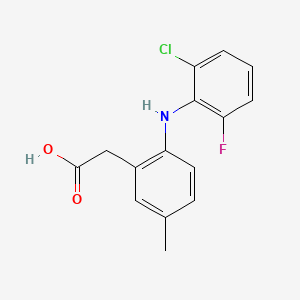 |
0.250 | ||
| ENC000068 | 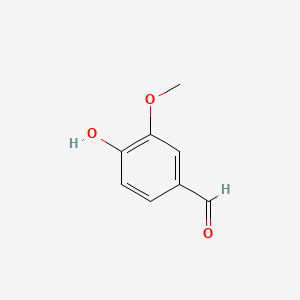 |
0.410 | D02WCI | 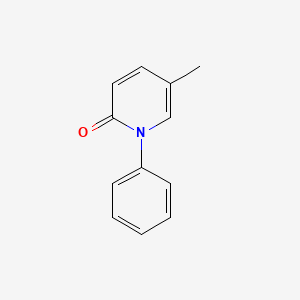 |
0.250 | ||
| ENC001334 | 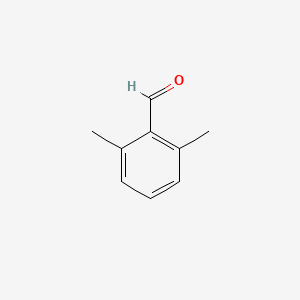 |
0.405 | D0T3NY | 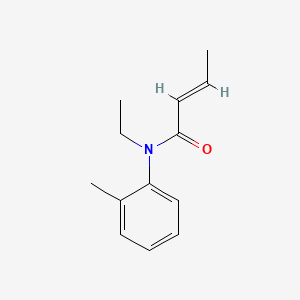 |
0.245 | ||
| ENC000005 | 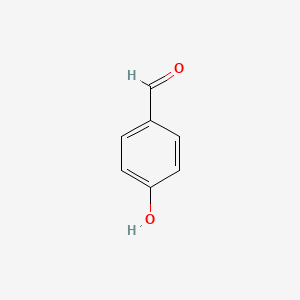 |
0.389 | D03GET |  |
0.245 | ||