NPs Basic Information
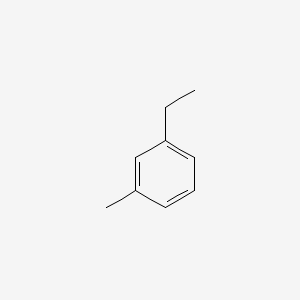
|
Name |
3-Ethyltoluene
|
| Molecular Formula | C9H12 | |
| IUPAC Name* |
1-ethyl-3-methylbenzene
|
|
| SMILES |
CCC1=CC=CC(=C1)C
|
|
| InChI |
InChI=1S/C9H12/c1-3-9-6-4-5-8(2)7-9/h4-7H,3H2,1-2H3
|
|
| InChIKey |
ZLCSFXXPPANWQY-UHFFFAOYSA-N
|
|
| Synonyms |
3-Ethyltoluene; 1-Ethyl-3-methylbenzene; 620-14-4; M-ETHYLTOLUENE; Benzene, 1-ethyl-3-methyl-; 1-Methyl-3-ethylbenzene; m-Ethylmethylbenzene; m-Methylethylbenzene; Toluene, m-ethyl-; 3-Methylethylbenzene; NSC 74176; 1-Ethyl-3-methyl-benzene; CHEMBL31274; 737PTD7O7E; CHEBI:77512; NSC-74176; 3-Ethyltoluene 100 microg/mL in Methanol; 3-ethyl-1-methylbenzene; EINECS 210-626-8; UNII-737PTD7O7E; m-Ethyl_toluene; meta-Ethyltoluene; 3-ethyl toluene; 3-ethylmethylbenzene; 1,3-methylethylbenzene; 3-Ethyltoluene, 97%; 3-Ethyltoluene, 99%; ETHYLTOLUENE, M-; DSSTox_CID_7876; DSSTox_RID_83511; DSSTox_GSID_50386; Benzene, 3-ethyl-1-methyl-; BIDD:ER0585; DTXSID6050386; ACT07964; NSC74176; ZINC1699560; Tox21_202857; BBL103656; BDBM50167946; MFCD00009259; STL557466; AKOS009158576; CS-W013572; NCGC00260403-01; CAS-620-14-4; DB-054039; E0185; FT-0615669; EN300-32024; F11740; A868581; Q27105073
|
|
| CAS | 620-14-4 | |
| PubChem CID | 12100 | |
| ChEMBL ID | CHEMBL31274 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 120.19 | ALogp: | 3.6 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.533 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.221 | MDCK Permeability: | 0.00002200 |
| Pgp-inhibitor: | 0.005 | Pgp-substrate: | 0.006 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.205 |
| 30% Bioavailability (F30%): | 0.558 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.945 | Plasma Protein Binding (PPB): | 91.41% |
| Volume Distribution (VD): | 2.098 | Fu: | 6.92% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.951 | CYP1A2-substrate: | 0.929 |
| CYP2C19-inhibitor: | 0.879 | CYP2C19-substrate: | 0.753 |
| CYP2C9-inhibitor: | 0.385 | CYP2C9-substrate: | 0.323 |
| CYP2D6-inhibitor: | 0.597 | CYP2D6-substrate: | 0.605 |
| CYP3A4-inhibitor: | 0.102 | CYP3A4-substrate: | 0.454 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.935 | Half-life (T1/2): | 0.661 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.048 | Human Hepatotoxicity (H-HT): | 0.094 |
| Drug-inuced Liver Injury (DILI): | 0.068 | AMES Toxicity: | 0.016 |
| Rat Oral Acute Toxicity: | 0.031 | Maximum Recommended Daily Dose: | 0.051 |
| Skin Sensitization: | 0.347 | Carcinogencity: | 0.245 |
| Eye Corrosion: | 0.978 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.082 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000612 |  |
0.571 | D0S5LH |  |
0.357 | ||
| ENC000239 |  |
0.567 | D02JIS | 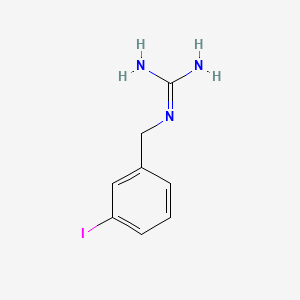 |
0.318 | ||
| ENC000368 | 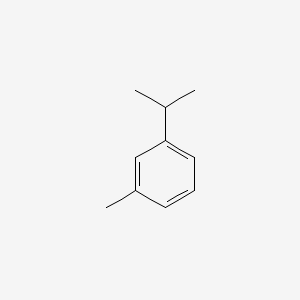 |
0.486 | D06GIP | 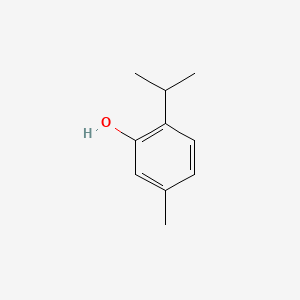 |
0.317 | ||
| ENC000414 | 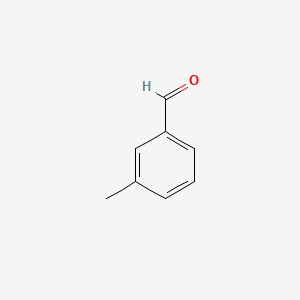 |
0.471 | D0U0RZ | 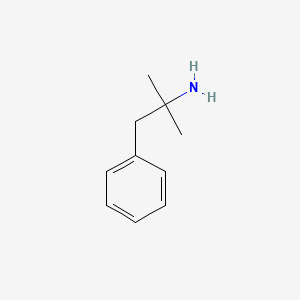 |
0.310 | ||
| ENC000203 |  |
0.455 | D0P6UB | 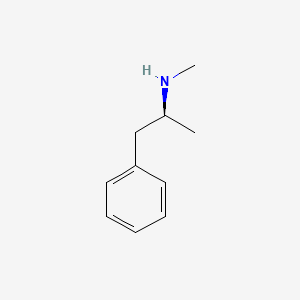 |
0.302 | ||
| ENC000370 | 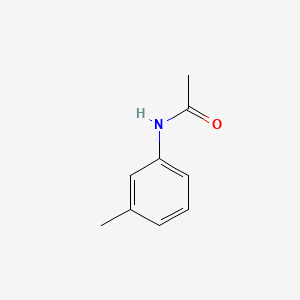 |
0.447 | D05OIS | 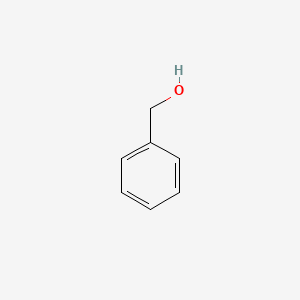 |
0.297 | ||
| ENC000734 | 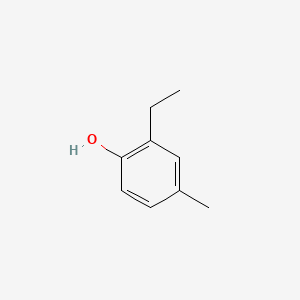 |
0.444 | D0T3LF | 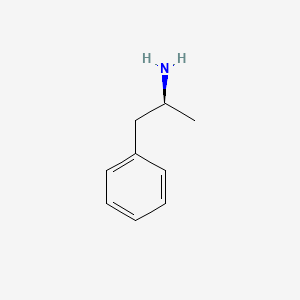 |
0.293 | ||
| ENC000498 | 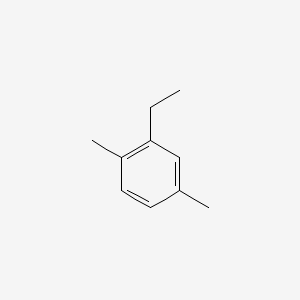 |
0.444 | D05BMG | 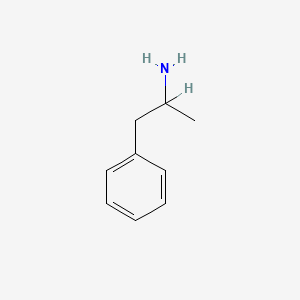 |
0.293 | ||
| ENC000407 |  |
0.429 | D0G1OZ |  |
0.289 | ||
| ENC000222 |  |
0.395 | D0O6IU |  |
0.289 | ||