NPs Basic Information
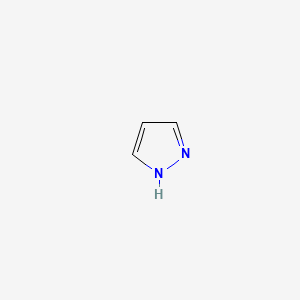
|
Name |
Pyrazole
|
| Molecular Formula | C3H4N2 | |
| IUPAC Name* |
1H-pyrazole
|
|
| SMILES |
C1=CNN=C1
|
|
| InChI |
InChI=1S/C3H4N2/c1-2-4-5-3-1/h1-3H,(H,4,5)
|
|
| InChIKey |
WTKZEGDFNFYCGP-UHFFFAOYSA-N
|
|
| Synonyms |
pyrazole; 1H-Pyrazole; 288-13-1; 1,2-diazole; 1H-pyrazol; Pyrazol; 3QD5KJZ7ZJ; CHEBI:17241; NSC45410; NSC-45410; 105809-46-9; Hpz; EINECS 206-017-1; UNII-3QD5KJZ7ZJ; MFCD00005234; NSC 45410; AI3-60151; Pyrazole-; 3-pyrazole; 1-h-pyrazole; Pyrazole, 98%; PYRAZOLE [MI]; WLN: T5MNJ; EC 206-017-1; CHEMBL15967; AMY795; DTXSID2059774; ZINC895257; ACT08445; BCP26863; STR00103; BDBM50390969; s3093; STK400566; AKOS000121045; Pyrazole, purum, >=98.0% (GC); CS-W008829; DB02757; BP-12835; HY-76228; NCI60_004054; Pyrazole, Vetec(TM) reagent grade, 98%; FT-0658357; P0546; EN300-21658; C00481; D78528; P-8110; 288P131; AC-907/25014006; Q408908; F2190-0642; Z104507728
|
|
| CAS | 288-13-1 | |
| PubChem CID | 1048 | |
| ChEMBL ID | CHEMBL15967 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 68.08 | ALogp: | 0.3 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 28.7 | Aromatic Rings: | 1 |
| Heavy Atoms: | 5 | QED Weighted: | 0.478 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.284 | MDCK Permeability: | 0.00002650 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.013 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.039 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.896 | Plasma Protein Binding (PPB): | 5.42% |
| Volume Distribution (VD): | 0.911 | Fu: | 88.47% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.062 | CYP1A2-substrate: | 0.614 |
| CYP2C19-inhibitor: | 0.075 | CYP2C19-substrate: | 0.164 |
| CYP2C9-inhibitor: | 0.015 | CYP2C9-substrate: | 0.865 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.479 |
| CYP3A4-inhibitor: | 0.013 | CYP3A4-substrate: | 0.118 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.271 | Half-life (T1/2): | 0.826 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.036 | Human Hepatotoxicity (H-HT): | 0.155 |
| Drug-inuced Liver Injury (DILI): | 0.951 | AMES Toxicity: | 0.013 |
| Rat Oral Acute Toxicity: | 0.034 | Maximum Recommended Daily Dose: | 0.047 |
| Skin Sensitization: | 0.537 | Carcinogencity: | 0.086 |
| Eye Corrosion: | 0.018 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.211 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000322 | 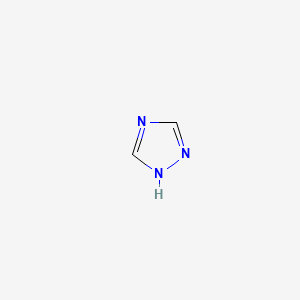 |
0.250 | D02NJA | 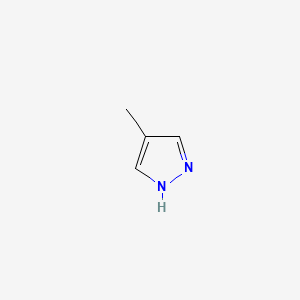 |
0.231 | ||
| ENC000440 | 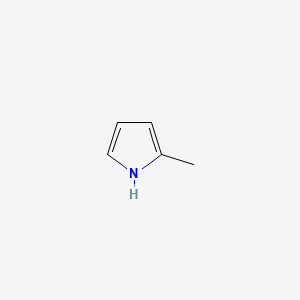 |
0.231 | D01OUE | 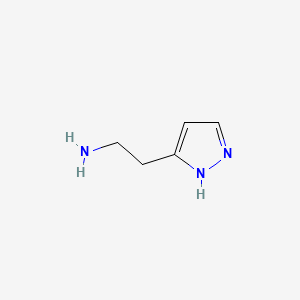 |
0.188 | ||
| ENC000439 | 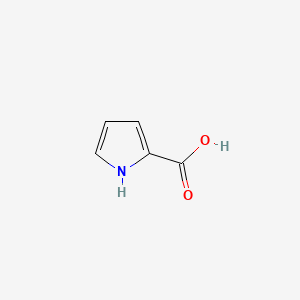 |
0.194 | D06NVJ | 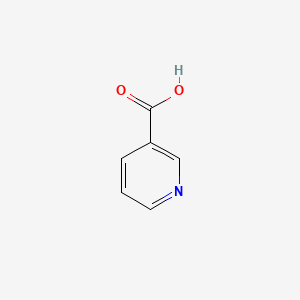 |
0.176 | ||
| ENC000056 | 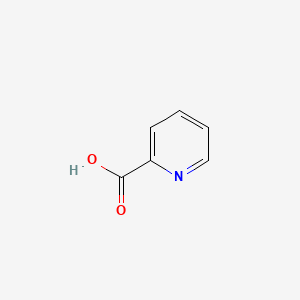 |
0.176 | D08YIN | 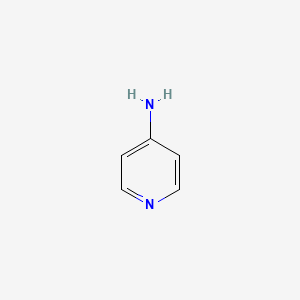 |
0.167 | ||
| ENC000485 | 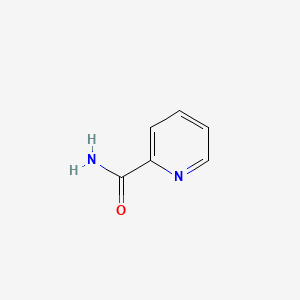 |
0.176 | D0F5ZM | 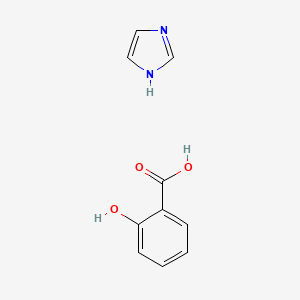 |
0.163 | ||
| ENC000048 | 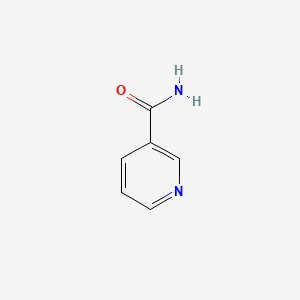 |
0.176 | D0XF8W |  |
0.143 | ||
| ENC000041 |  |
0.167 | D05QIM | 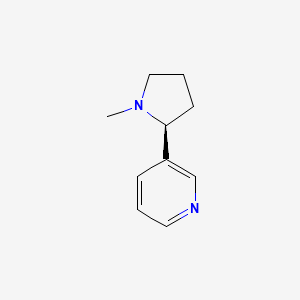 |
0.136 | ||
| ENC000240 |  |
0.156 | D0S4BR | 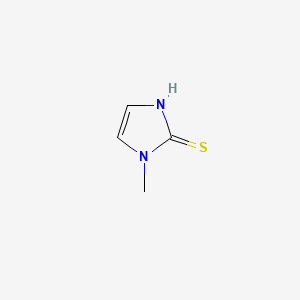 |
0.133 | ||
| ENC001061 | 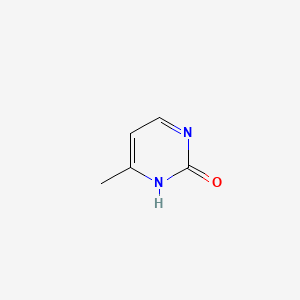 |
0.156 | D03RZV | 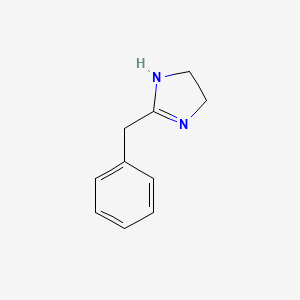 |
0.133 | ||
| ENC000663 | 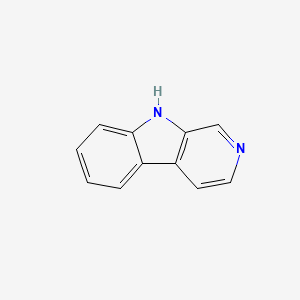 |
0.149 | D06AEB | 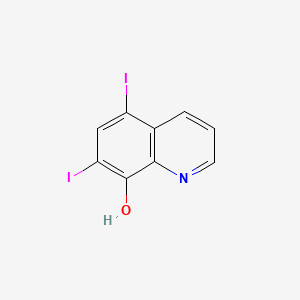 |
0.133 | ||