NPs Basic Information
|
Name |
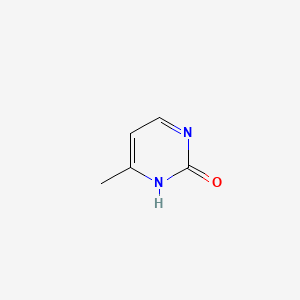
2-Hydroxy-4-methylpyrimidine
|
| Molecular Formula | C5H6N2O | |
| IUPAC Name* |
6-methyl-1H-pyrimidin-2-one
|
|
| SMILES |
CC1=CC=NC(=O)N1
|
|
| InChI |
InChI=1S/C5H6N2O/c1-4-2-3-6-5(8)7-4/h2-3H,1H3,(H,6,7,8)
|
|
| InChIKey |
AHHHDTLXONDKQF-UHFFFAOYSA-N
|
|
| Synonyms |
4-methylpyrimidin-2-ol; 2-Hydroxy-4-methylpyrimidine; 15231-48-8; 4-methyl-1,2-dihydropyrimidin-2-one; 4-METHYLPYRIMIDIN-2(1H)-ONE; 6-methyl-1H-pyrimidin-2-one; 4-methyl-2-pyrimidinol; 4-Methyl-2(1H)-pyrimidinone; 6-Methylpyrimidin-2(1H)-one; 15231-67-1; 2(1H)-Pyrimidinone, 4-methyl-; 4-methylpyrimid-2-one; 6-Methyl-2-pyrimidone; 6-methyl-2-pyrimidinone; 2PYRIMIDONE4METHYL; 4-Methyl-pyrimidin-2-ol; hydroxy-6-methylpyrimidine; 2-Pyrimidinol, 4-methyl-; 4-Methylpyrimidin-2-ol HCl; 4-Methyl-2-hydroxypyrimidine; SCHEMBL501750; 4-Methyl-5H-pyrimidin-2-one; 2(1H)-Pyrimidinone,4-methyl-; 4-METHYLPYRIMIDIN-2-ONE; SCHEMBL10325788; AHHHDTLXONDKQF-UHFFFAOYSA-; DTXSID70878778; 4-Methyl-2-oxo-(1H)-pyrimidine; ACT08959; ALBB-016443; NSC 1588; EINECS 226-306-6; MFCD00044500; MFCD09991726; ZINC18084478; AKOS002337540; AKOS006230604; PB19074; 2-pyrimidinol, 4-methyl-, hydrochloride; AM803253; AS-50692; SY073173; 4-methyl-3H-pyrimidin-2-one Hydrochloride; DB-081949; CS-0037723; FT-0648931; EN300-66580; P10883; W10295; AC-907/30003036; W-201357; Z1065584884
|
|
| CAS | 5348-51-6 | |
| PubChem CID | 407091 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 110.11 | ALogp: | -1.4 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 41.5 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.528 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.466 | MDCK Permeability: | 0.00001850 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.016 |
| Human Intestinal Absorption (HIA): | 0.013 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.604 | Plasma Protein Binding (PPB): | 33.03% |
| Volume Distribution (VD): | 0.839 | Fu: | 68.06% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.034 | CYP1A2-substrate: | 0.906 |
| CYP2C19-inhibitor: | 0.046 | CYP2C19-substrate: | 0.061 |
| CYP2C9-inhibitor: | 0.008 | CYP2C9-substrate: | 0.243 |
| CYP2D6-inhibitor: | 0.015 | CYP2D6-substrate: | 0.155 |
| CYP3A4-inhibitor: | 0.006 | CYP3A4-substrate: | 0.18 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.341 | Half-life (T1/2): | 0.826 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.709 |
| Drug-inuced Liver Injury (DILI): | 0.949 | AMES Toxicity: | 0.02 |
| Rat Oral Acute Toxicity: | 0.121 | Maximum Recommended Daily Dose: | 0.026 |
| Skin Sensitization: | 0.408 | Carcinogencity: | 0.782 |
| Eye Corrosion: | 0.022 | Eye Irritation: | 0.959 |
| Respiratory Toxicity: | 0.052 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000721 | 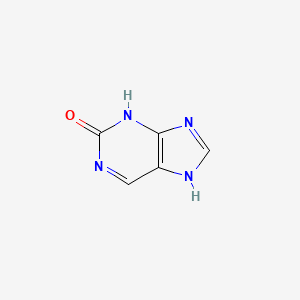 |
0.308 | D0S5WG | 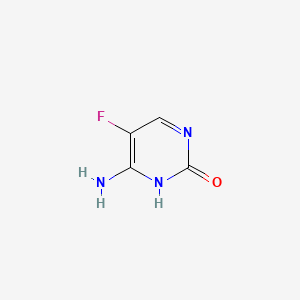 |
0.314 | ||
| ENC000440 | 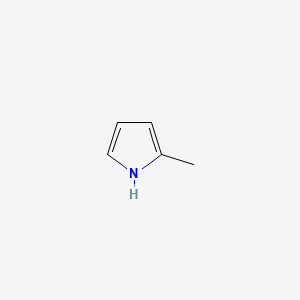 |
0.258 | D0Y9ZE | 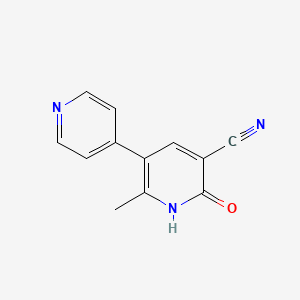 |
0.241 | ||
| ENC000065 | 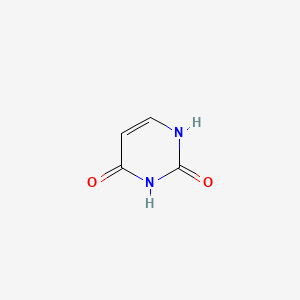 |
0.257 | D0P0HB | 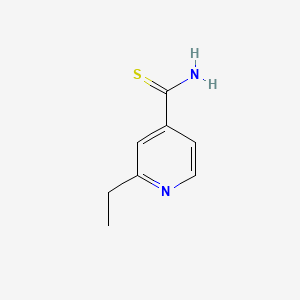 |
0.238 | ||
| ENC000178 | 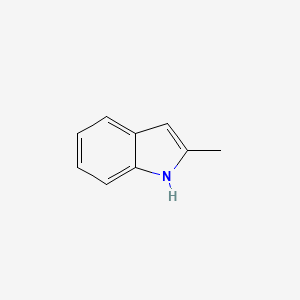 |
0.244 | D0N0OU | 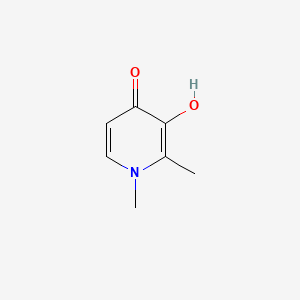 |
0.231 | ||
| ENC003235 | 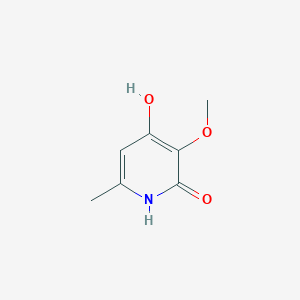 |
0.244 | D03ZBN | 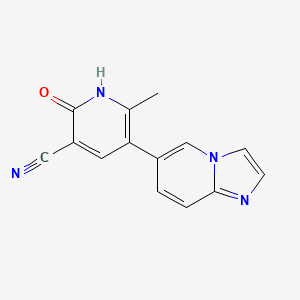 |
0.226 | ||
| ENC000063 | 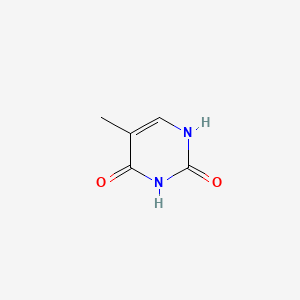 |
0.243 | D0L7UQ | 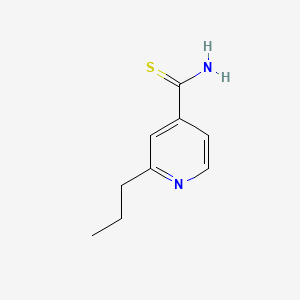 |
0.222 | ||
| ENC000292 | 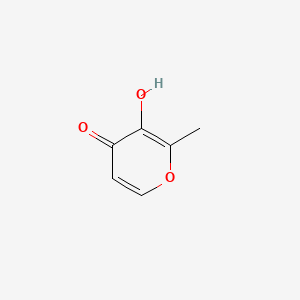 |
0.243 | D02NJA | 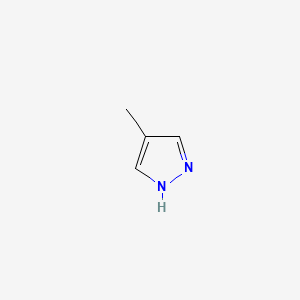 |
0.219 | ||
| ENC000577 | 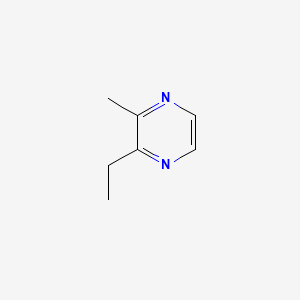 |
0.237 | D0O2EM | 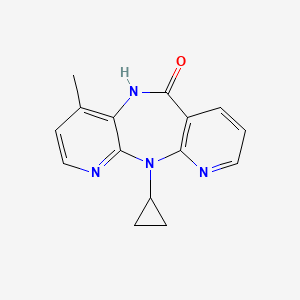 |
0.215 | ||
| ENC005053 | 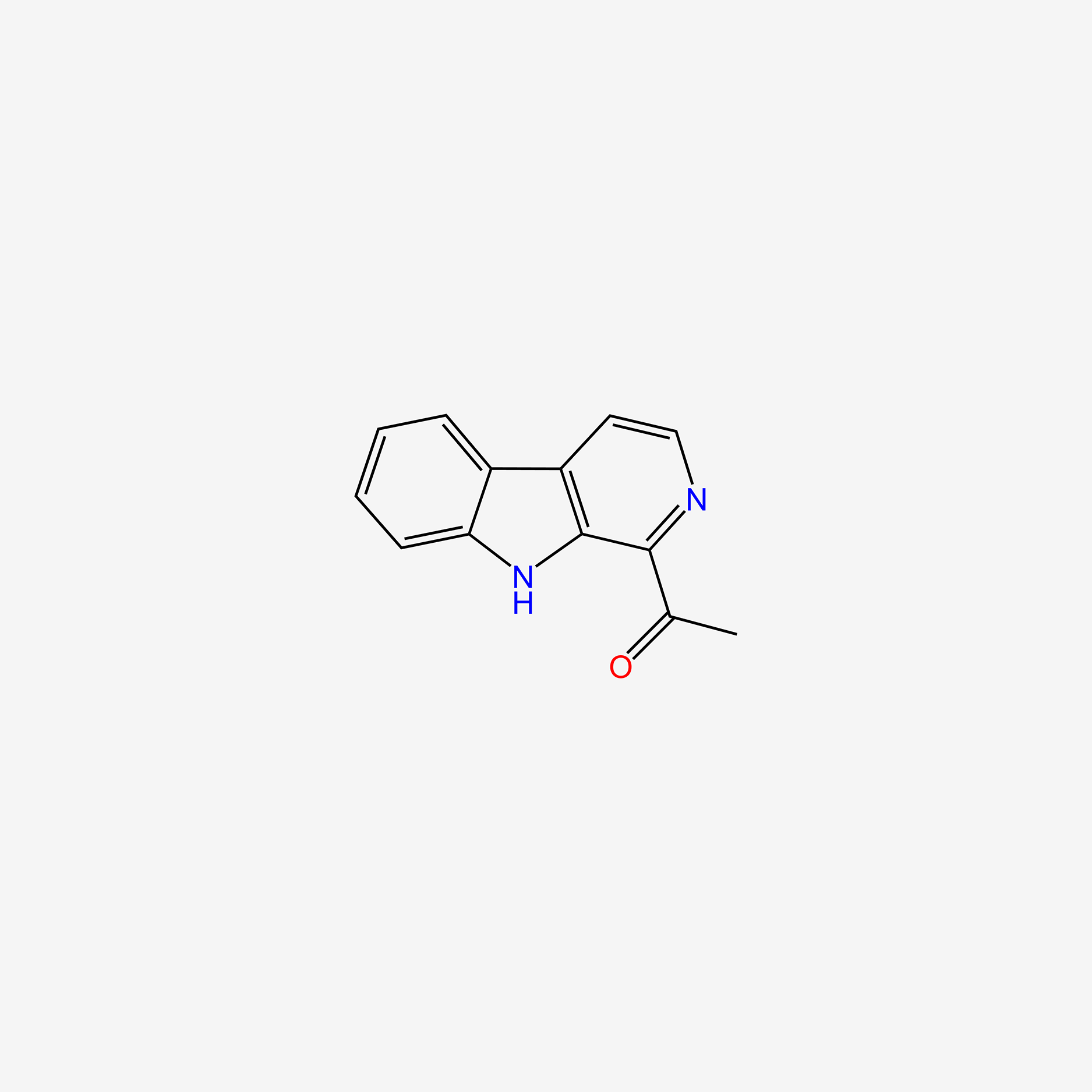 |
0.236 | D04KYY | 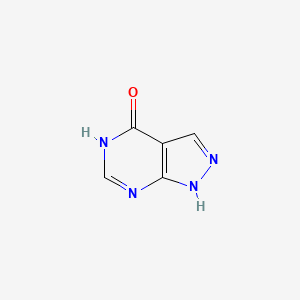 |
0.214 | ||
| ENC001342 | 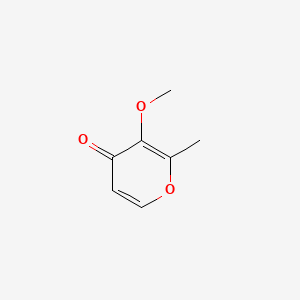 |
0.225 | D0S1NZ | 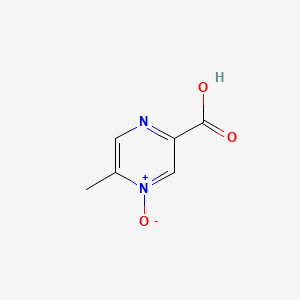 |
0.214 | ||