NPs Basic Information
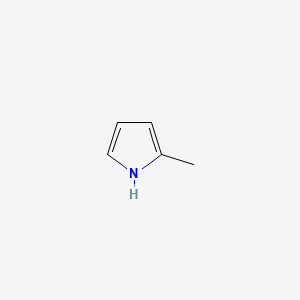
|
Name |
2-Methyl-1H-pyrrole
|
| Molecular Formula | C5H7N | |
| IUPAC Name* |
2-methyl-1H-pyrrole
|
|
| SMILES |
CC1=CC=CN1
|
|
| InChI |
InChI=1S/C5H7N/c1-5-3-2-4-6-5/h2-4,6H,1H3
|
|
| InChIKey |
TVCXVUHHCUYLGX-UHFFFAOYSA-N
|
|
| Synonyms |
2-Methyl-1H-pyrrole; 636-41-9; 2-METHYLPYRROLE; 1H-Pyrrole, 2-methyl-; Pyrrole, 2-methyl-; alpha-Methylpyrrole; 2-Methylpyrolle; .alpha.-Methylpyrrole; MFCD02822910; NSC-81346; 486RY4814O; a-Methylpyrrole; UNII-486RY4814O; 2-Methyl pyrrole; 2-Methyl-Pyrrole; (2-pyrryl)methane; NSC81346; alpha -methylpyrrole; EINECS 211-255-4; NSC 81346; 2-methyl-1H-pyrazole; 2-Methyl-1H-pyrrole #; Pyrrole, 2-methyl- (8CI); DTXSID70212976; CHEBI:193616; 1H-Pyrrole, 2-methyl- (9CI); AMY18997; ZINC1574399; AKOS006221233; AB12705; CS-W011360; DA-25958; SY004787; FT-0762789; EN300-49636; 93M981; A868079; Q27259111; Z594218440
|
|
| CAS | 636-41-9 | |
| PubChem CID | 12489 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 81.12 | ALogp: | 1.1 |
| HBD: | 1 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 15.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 6 | QED Weighted: | 0.49 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.22 | MDCK Permeability: | 0.00002760 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.128 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.009 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.948 | Plasma Protein Binding (PPB): | 48.01% |
| Volume Distribution (VD): | 2.103 | Fu: | 63.35% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.55 | CYP1A2-substrate: | 0.678 |
| CYP2C19-inhibitor: | 0.236 | CYP2C19-substrate: | 0.459 |
| CYP2C9-inhibitor: | 0.024 | CYP2C9-substrate: | 0.764 |
| CYP2D6-inhibitor: | 0.168 | CYP2D6-substrate: | 0.706 |
| CYP3A4-inhibitor: | 0.019 | CYP3A4-substrate: | 0.207 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.247 | Half-life (T1/2): | 0.847 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.029 | Human Hepatotoxicity (H-HT): | 0.391 |
| Drug-inuced Liver Injury (DILI): | 0.086 | AMES Toxicity: | 0.056 |
| Rat Oral Acute Toxicity: | 0.949 | Maximum Recommended Daily Dose: | 0.07 |
| Skin Sensitization: | 0.625 | Carcinogencity: | 0.151 |
| Eye Corrosion: | 0.953 | Eye Irritation: | 0.993 |
| Respiratory Toxicity: | 0.972 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000439 | 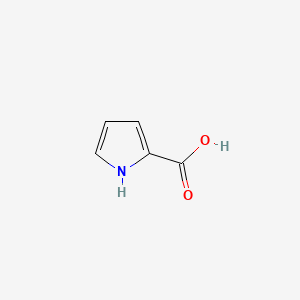 |
0.393 | D0S4BR | 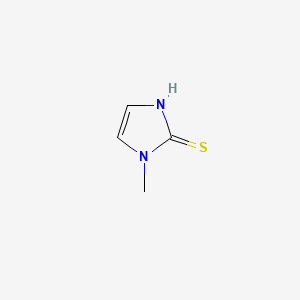 |
0.241 | ||
| ENC005078 | 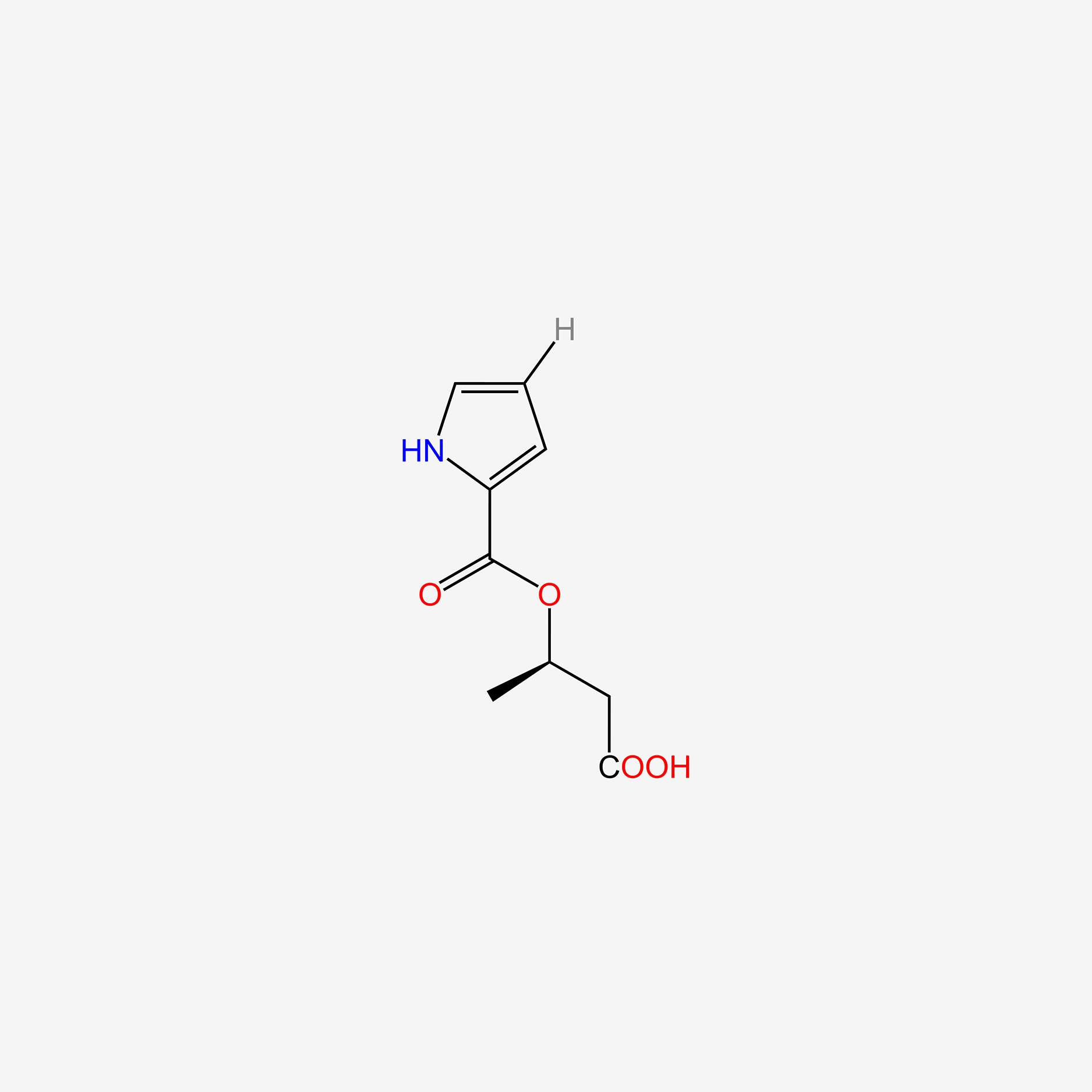 |
0.279 | D02NJA | 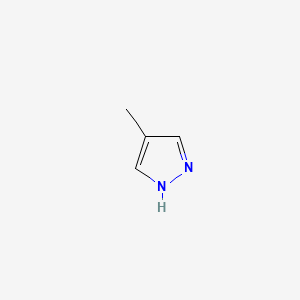 |
0.214 | ||
| ENC000178 | 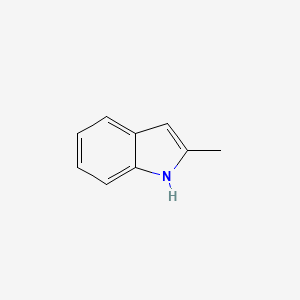 |
0.278 | D0U3DU | 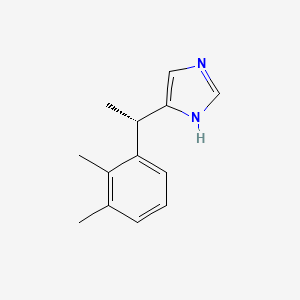 |
0.204 | ||
| ENC000064 | 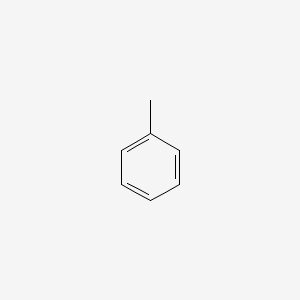 |
0.276 | D0T3LF | 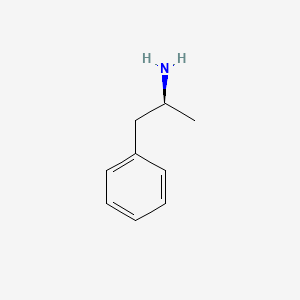 |
0.184 | ||
| ENC000028 | 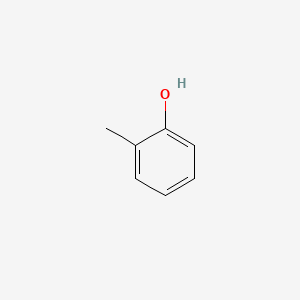 |
0.258 | D05BMG | 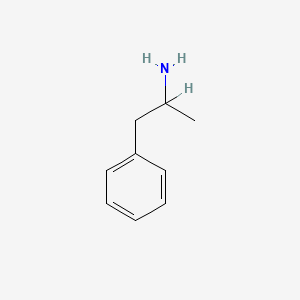 |
0.184 | ||
| ENC001061 | 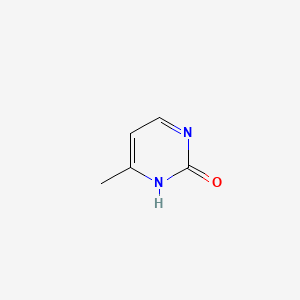 |
0.258 | D0X0RI | 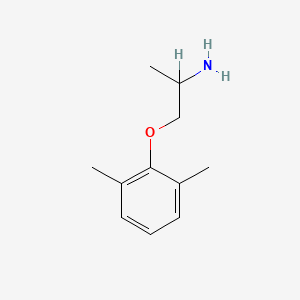 |
0.182 | ||
| ENC000179 | 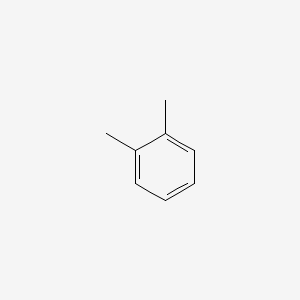 |
0.258 | D06GIP | 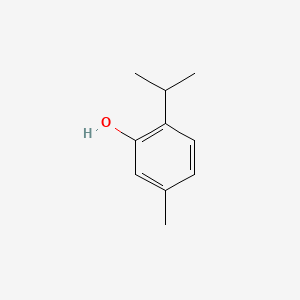 |
0.179 | ||
| ENC000240 |  |
0.258 | D0X7NU | 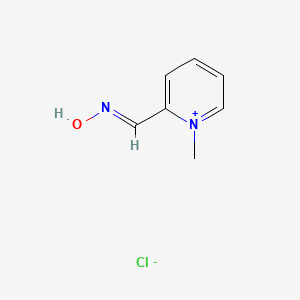 |
0.179 | ||
| ENC000239 | 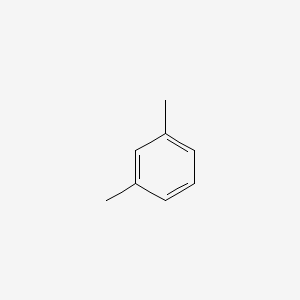 |
0.258 | D03GET | 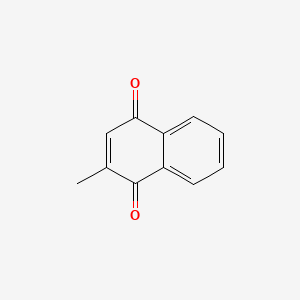 |
0.178 | ||
| ENC000041 |  |
0.257 | D05OIS | 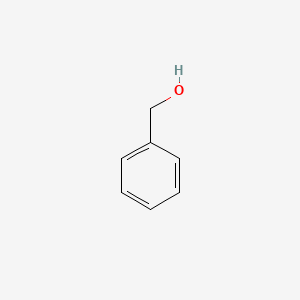 |
0.176 | ||