NPs Basic Information
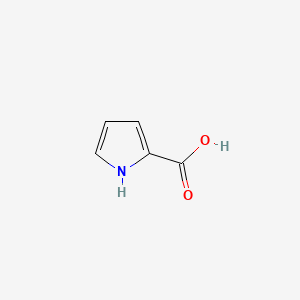
|
Name |
Pyrrole-2-carboxylic acid
|
| Molecular Formula | C5H5NO2 | |
| IUPAC Name* |
1H-pyrrole-2-carboxylic acid
|
|
| SMILES |
C1=CNC(=C1)C(=O)O
|
|
| InChI |
InChI=1S/C5H5NO2/c7-5(8)4-2-1-3-6-4/h1-3,6H,(H,7,8)
|
|
| InChIKey |
WRHZVMBBRYBTKZ-UHFFFAOYSA-N
|
|
| Synonyms |
PYRROLE-2-CARBOXYLIC ACID; 634-97-9; 1H-Pyrrole-2-carboxylic acid; Minaline; 2-Pyrrolecarboxylic acid; Minalin; Pyrrole-2-carboxylicacid; Pyrrole-2-carboxylate; 1H-Pyrrolecarboxylic acid; MFCD00005219; 2NNB85QQT9; CHEMBL509027; CHEBI:36751; NSC-48130; PYC; Mialine; 2-Minaline; EINECS 211-221-9; NSC 48130; Pyrrole 2-carboxylate; Pyrrol-2-carboxylic acid; Pyrrole 2-carboxylic acid; 1H-pyrrole-carboxylic acid; UNII-2NNB85QQT9; SCHEMBL28298; SCHEMBL14725915; DTXSID50212813; Pyrrole-2-carboxylic acid, 99%; ZINC158648; ACT01693; BCP29322; NSC48130; STR04784; BDBM50260723; s5832; STL164370; 1H-Pyrrole-2-carboxylic acid (9CI); AKOS001132829; AC-6174; CS-W001963; DB02543; FG-0411; HY-W001963; PB47384; SY014890; Minaline pound>>Pyrrole-2-carboxylicacid; DB-011234; A8742; AM20100569; BB 0269481; FT-0632694; P1270; Pyrrole-2-carboxylic acid, Streptomyces sp.; EN300-13984; C05942; 634P979; AC-907/25014066; W-104908; Q27104383; Z94599900; 1H-pyrrole-2-carboxylic acid;Pyrrole-2-carboxylic acid; F8880-6911; 98C22043-F536-4DC6-8C61-12A0C5E6A5D6
|
|
| CAS | 634-97-9 | |
| PubChem CID | 12473 | |
| ChEMBL ID | CHEMBL509027 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 111.1 | ALogp: | 0.8 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 53.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.57 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.012 | MDCK Permeability: | 0.00000711 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.014 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.541 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.893 | Plasma Protein Binding (PPB): | 14.73% |
| Volume Distribution (VD): | 0.25 | Fu: | 76.90% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.024 | CYP1A2-substrate: | 0.098 |
| CYP2C19-inhibitor: | 0.052 | CYP2C19-substrate: | 0.05 |
| CYP2C9-inhibitor: | 0.021 | CYP2C9-substrate: | 0.406 |
| CYP2D6-inhibitor: | 0.008 | CYP2D6-substrate: | 0.141 |
| CYP3A4-inhibitor: | 0.015 | CYP3A4-substrate: | 0.045 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.809 | Half-life (T1/2): | 0.92 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.032 | Human Hepatotoxicity (H-HT): | 0.241 |
| Drug-inuced Liver Injury (DILI): | 0.715 | AMES Toxicity: | 0.015 |
| Rat Oral Acute Toxicity: | 0.941 | Maximum Recommended Daily Dose: | 0.008 |
| Skin Sensitization: | 0.152 | Carcinogencity: | 0.054 |
| Eye Corrosion: | 0.122 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.936 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005078 | 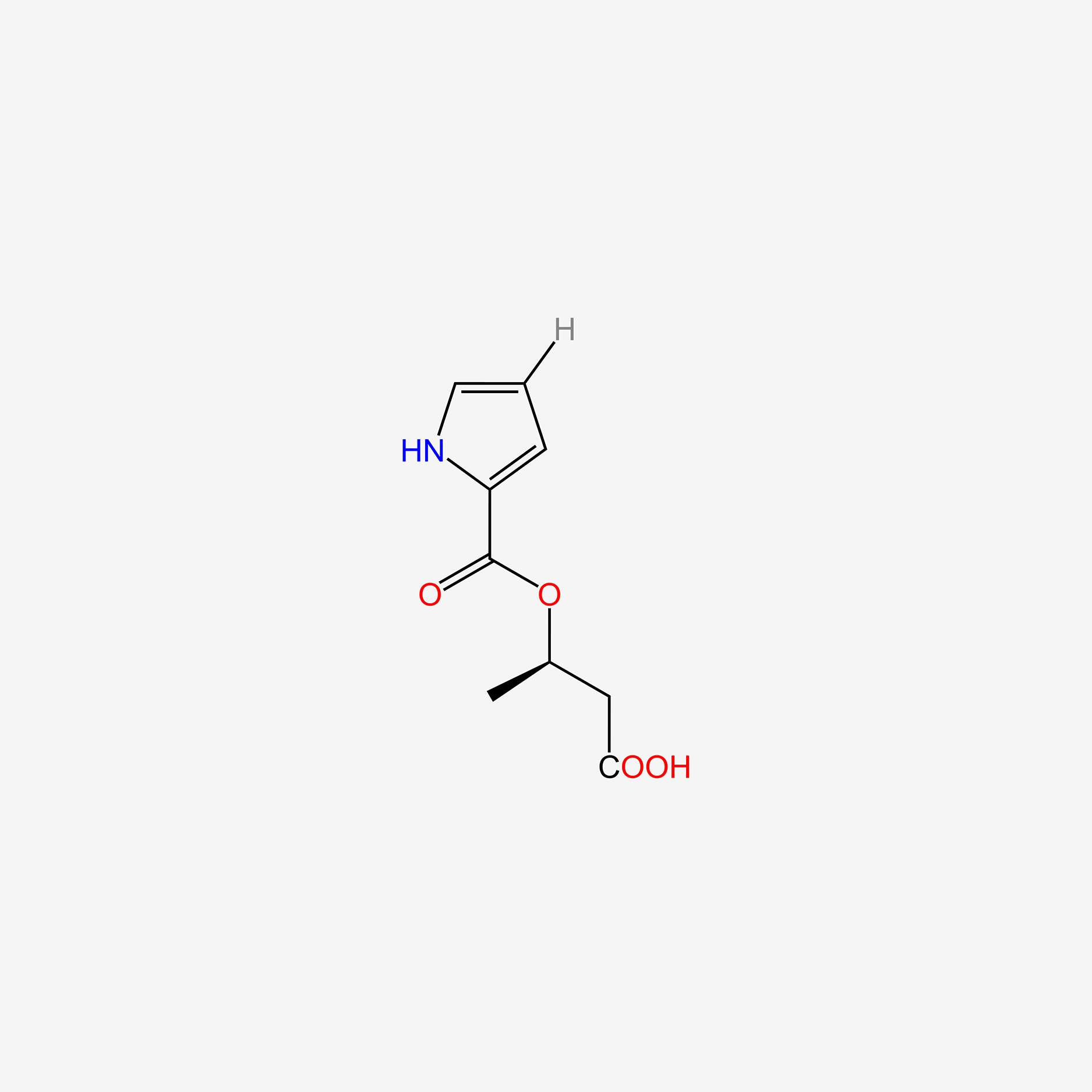 |
0.463 | D06NVJ | 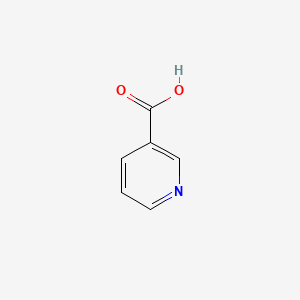 |
0.343 | ||
| ENC005079 | 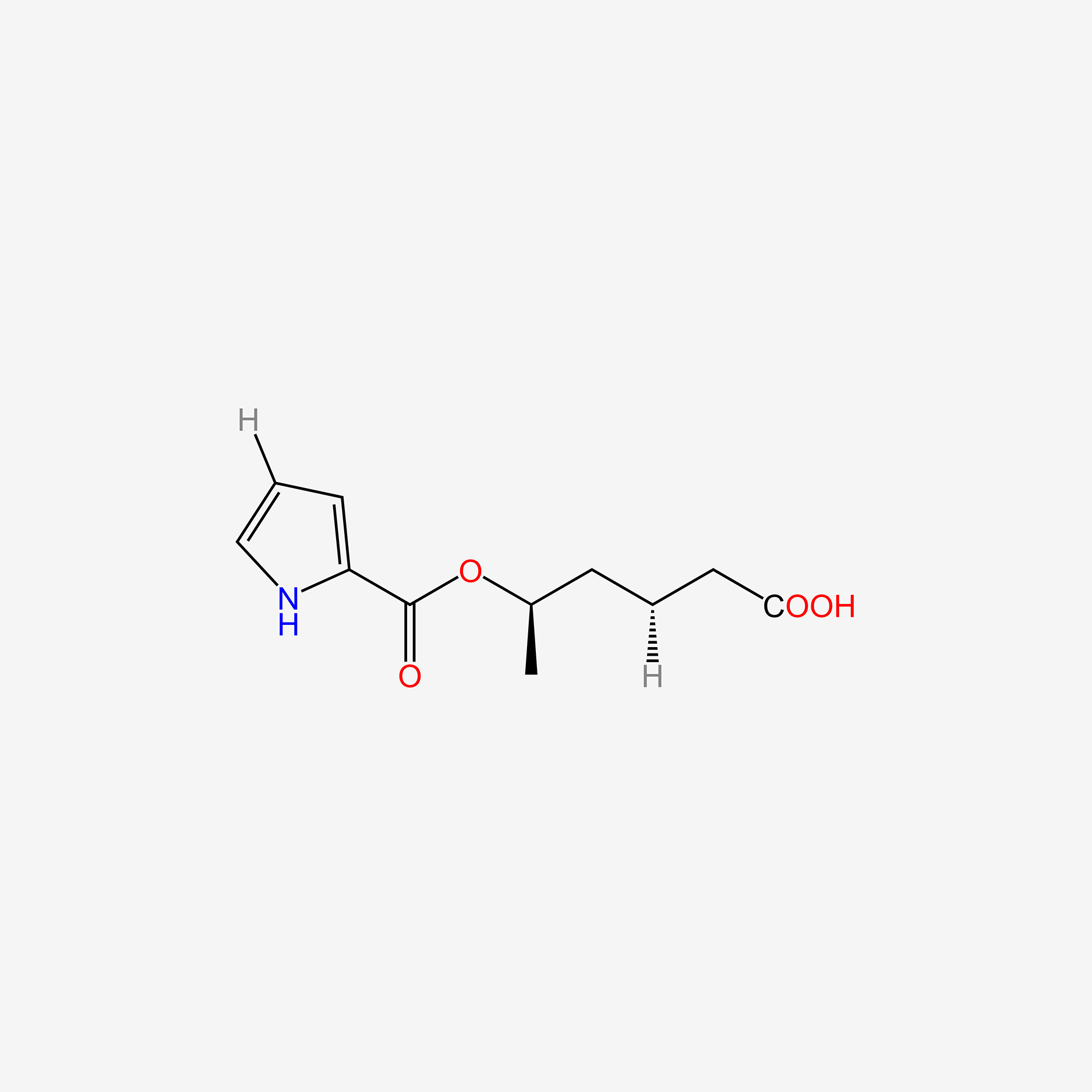 |
0.404 | D07HBX |  |
0.324 | ||
| ENC000440 | 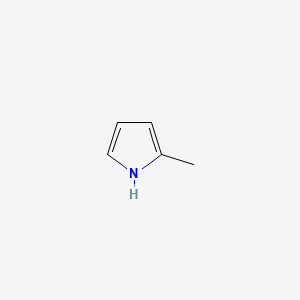 |
0.393 | D0F5ZM | 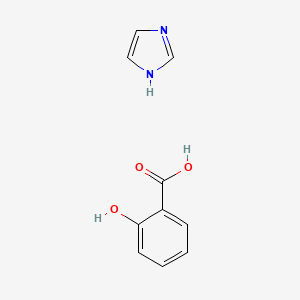 |
0.280 | ||
| ENC005083 | 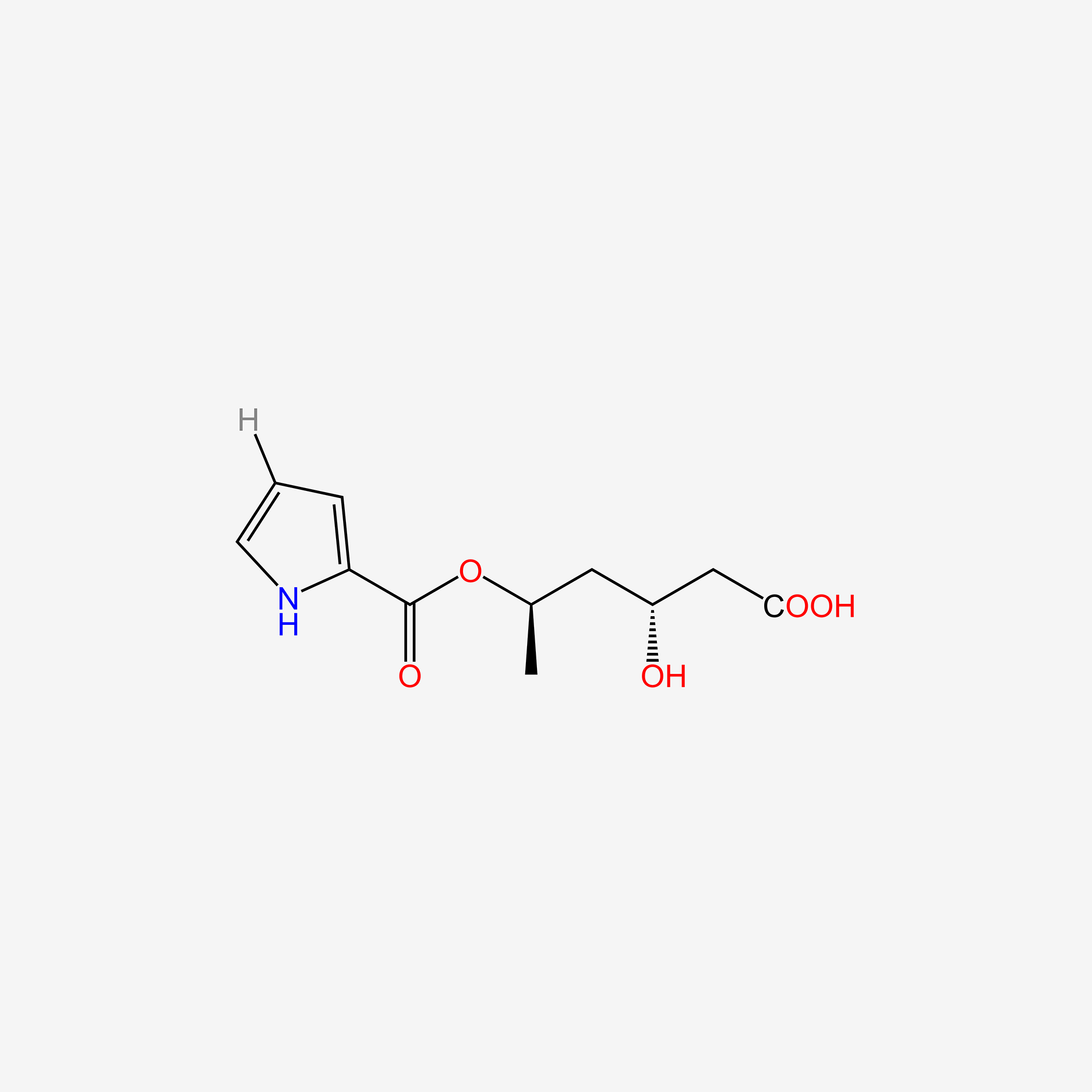 |
0.388 | D0C4YC | 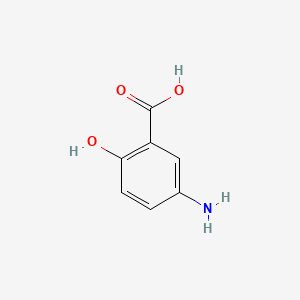 |
0.275 | ||
| ENC000162 | 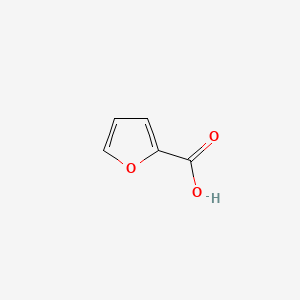 |
0.375 | D01WJL | 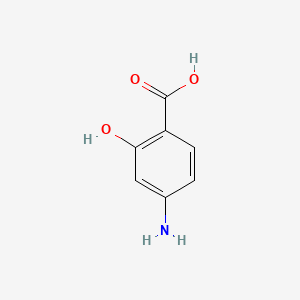 |
0.275 | ||
| ENC005084 | 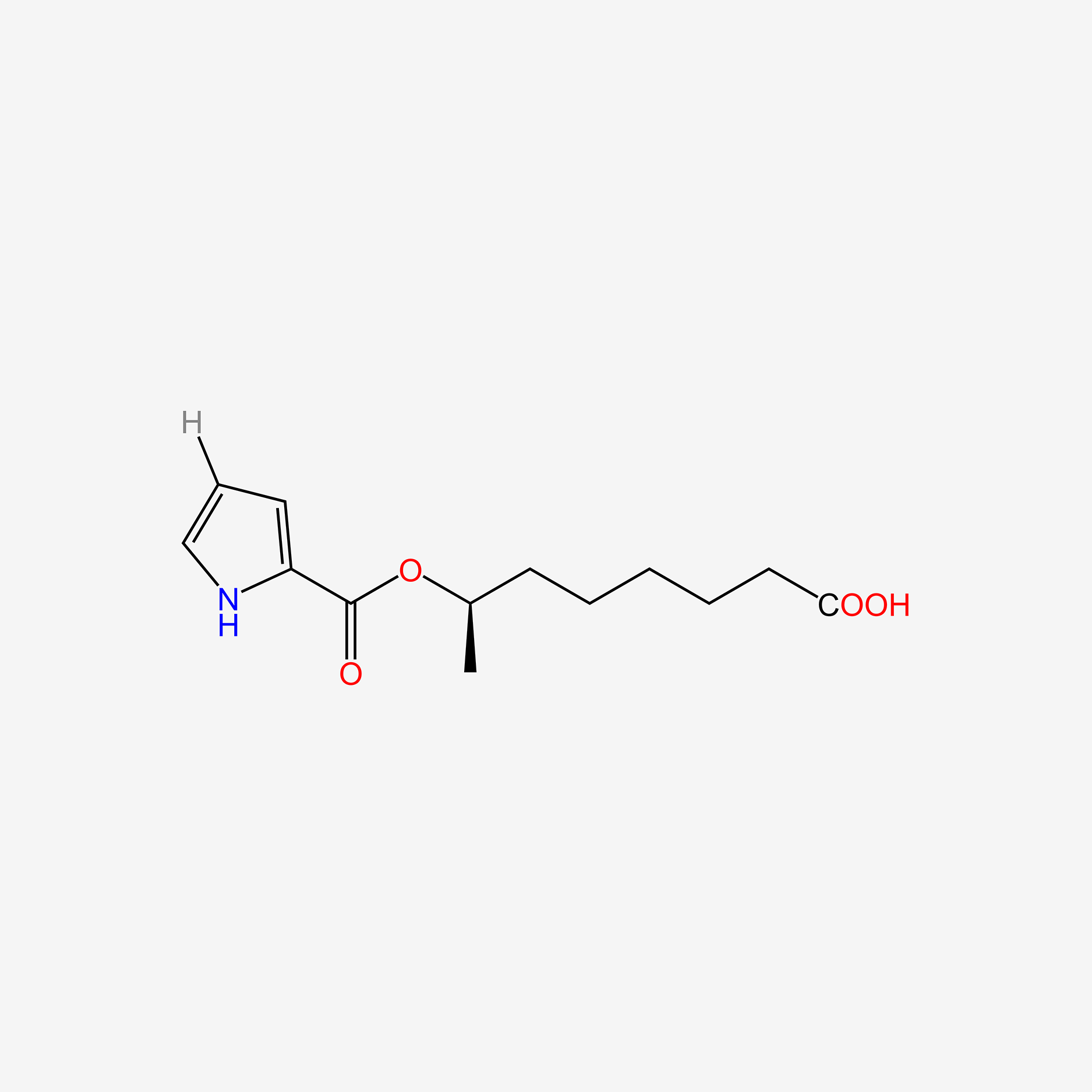 |
0.358 | D0GY5Z | 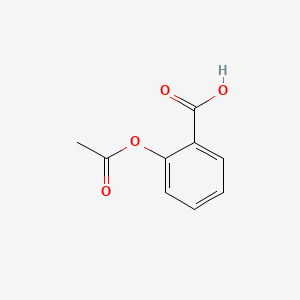 |
0.267 | ||
| ENC000013 | 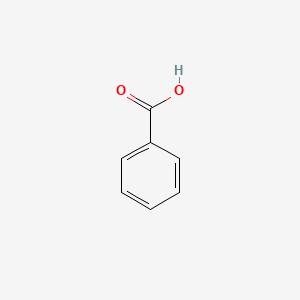 |
0.343 | D01ZJK | 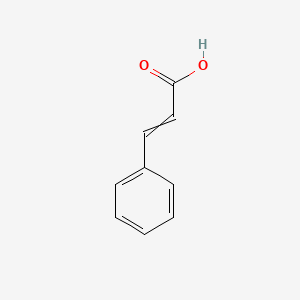 |
0.262 | ||
| ENC000056 | 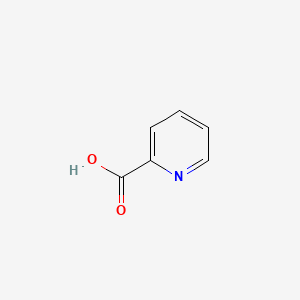 |
0.343 | D0N3UL | 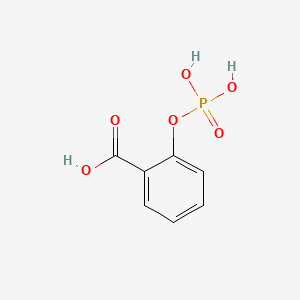 |
0.255 | ||
| ENC005757 | 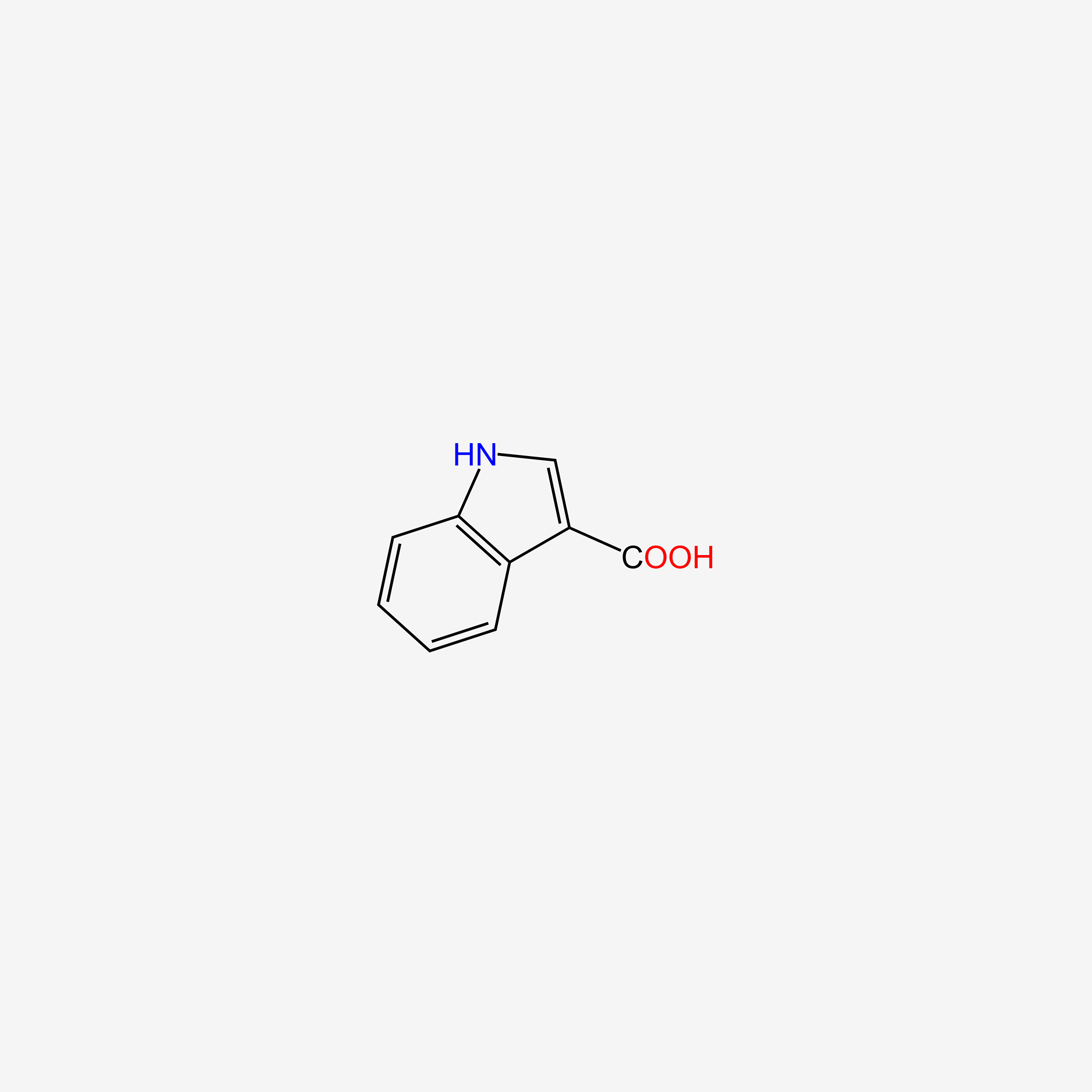 |
0.333 | D05EJG | 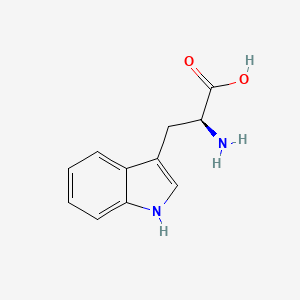 |
0.255 | ||
| ENC000118 | 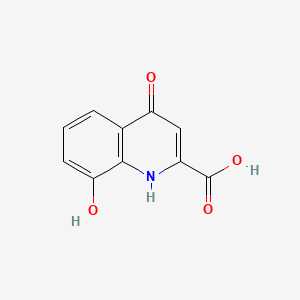 |
0.313 | D0R1CR | 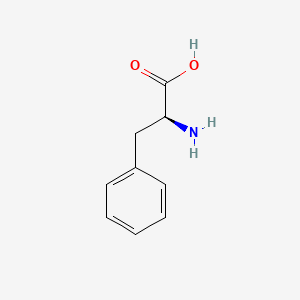 |
0.250 | ||