NPs Basic Information
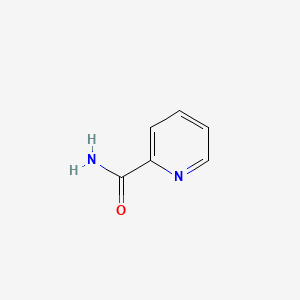
|
Name |
Picolinamide
|
| Molecular Formula | C6H6N2O | |
| IUPAC Name* |
pyridine-2-carboxamide
|
|
| SMILES |
C1=CC=NC(=C1)C(=O)N
|
|
| InChI |
InChI=1S/C6H6N2O/c7-6(9)5-3-1-2-4-8-5/h1-4H,(H2,7,9)
|
|
| InChIKey |
IBBMAWULFFBRKK-UHFFFAOYSA-N
|
|
| Synonyms |
Picolinamide; 1452-77-3; PYRIDINE-2-CARBOXAMIDE; 2-Pyridinecarboxamide; Picolinoylamide; 2-Picolinamide; 2-Carbamoylpyridine; Picolinic acid amide; 2-aminocarbonyl-pyridine; alpha-Picolinamide; Pyridinecarboxamide; .alpha.-Picolinic acid amide; I3550CCL59; alpha - picolinamide; MFCD00023483; Pyridine-2-carboxamide (Picolinamide); NSC-524473; picolamide; pyridine amide; UNII-I3550CCL59; 2-picoloylamine; pyridyl carboxamide; EINECS 215-921-5; .alpha.-Picolinamide; Picolinamide, 98%; NSC 524473; 2-pyridine-carboxamide; pyridine 2-carboxamide; 2-picolinic acid amide; 2-pyridinecarboximidic acid; .alpha.-Pyridinecarboxamide; PICOLINAMIDE [INCI]; SCHEMBL61183; CHEBI:8200; DTXSID4061703; SCHEMBL11253275; SCHEMBL11256944; IBBMAWULFFBRKK-UHFFFAOYSA-; pyridine-2-carboxylic acid amide; HMS1784A15; ZINC388048; Picolinamide; 2-Pyridinecarboxamide; NSC524473; s4710; AKOS001144712; AM85589; CCG-266081; GS-6464; SY048638; DB-042775; HY-101020; CS-0020700; FT-0622194; P0414; C01950; A808344; J-008082; Q27107933; Z33546380; 2-Pyridinecarboxamide; Picolinoylamide; 2-Carbamoylpyridine; WM1
|
|
| CAS | 1452-77-3 | |
| PubChem CID | 15070 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 122.12 | ALogp: | 0.2 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 56.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.592 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.336 | MDCK Permeability: | 0.00003240 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.022 |
| Human Intestinal Absorption (HIA): | 0.007 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.032 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.99 | Plasma Protein Binding (PPB): | 23.32% |
| Volume Distribution (VD): | 1.276 | Fu: | 77.83% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.249 | CYP1A2-substrate: | 0.332 |
| CYP2C19-inhibitor: | 0.029 | CYP2C19-substrate: | 0.061 |
| CYP2C9-inhibitor: | 0.008 | CYP2C9-substrate: | 0.172 |
| CYP2D6-inhibitor: | 0.004 | CYP2D6-substrate: | 0.381 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.173 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.043 | Half-life (T1/2): | 0.268 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.084 | Human Hepatotoxicity (H-HT): | 0.099 |
| Drug-inuced Liver Injury (DILI): | 0.909 | AMES Toxicity: | 0.019 |
| Rat Oral Acute Toxicity: | 0.471 | Maximum Recommended Daily Dose: | 0.019 |
| Skin Sensitization: | 0.127 | Carcinogencity: | 0.065 |
| Eye Corrosion: | 0.011 | Eye Irritation: | 0.983 |
| Respiratory Toxicity: | 0.506 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000056 | 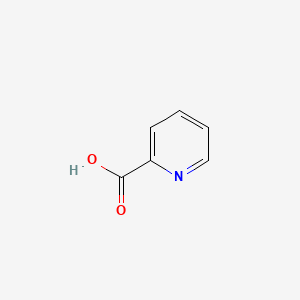 |
0.667 | D0XF8W |  |
0.471 | ||
| ENC000585 | 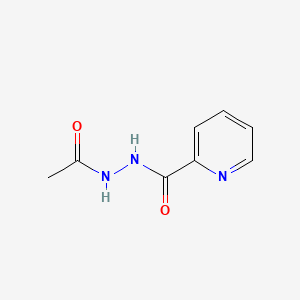 |
0.488 | D06NVJ | 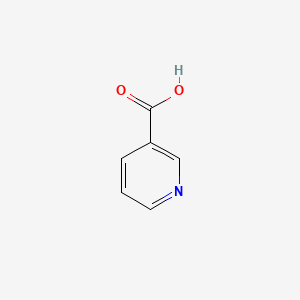 |
0.316 | ||
| ENC000048 | 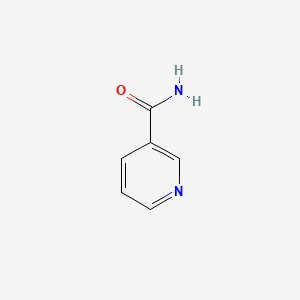 |
0.429 | D0X9RY | 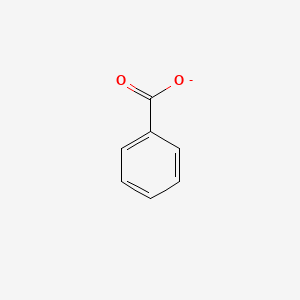 |
0.316 | ||
| ENC000076 | 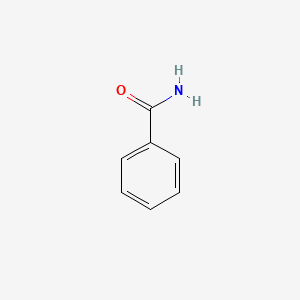 |
0.429 | D07HBX |  |
0.300 | ||
| ENC000108 | 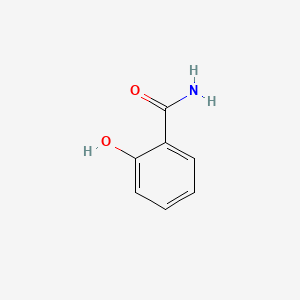 |
0.405 | D07ONP | 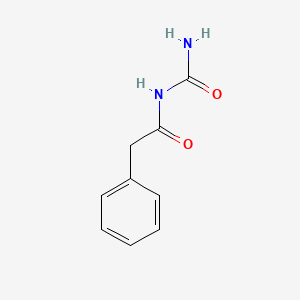 |
0.298 | ||
| ENC000219 | 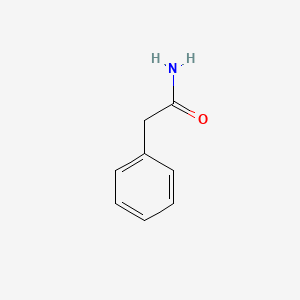 |
0.359 | D09XQF | 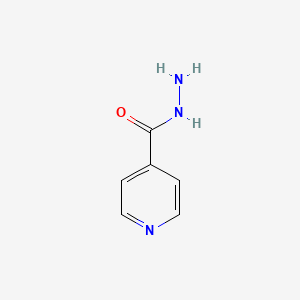 |
0.293 | ||
| ENC005854 |  |
0.359 | D0R1CR | 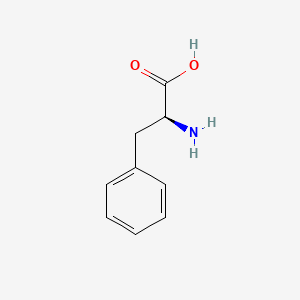 |
0.289 | ||
| ENC001049 | 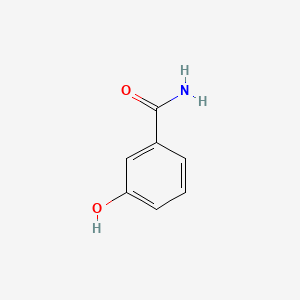 |
0.333 | D0MD2L |  |
0.286 | ||
| ENC000192 | 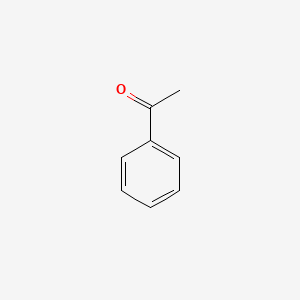 |
0.316 | D0D4CY | 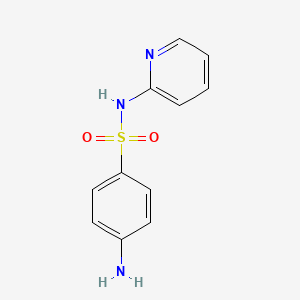 |
0.281 | ||
| ENC000013 | 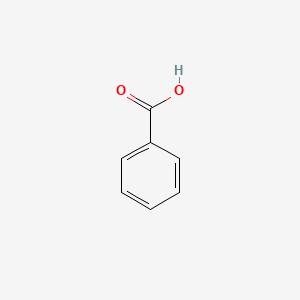 |
0.316 | D01ZJK |  |
0.273 | ||