NPs Basic Information
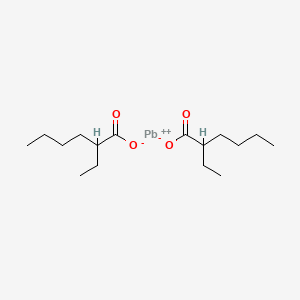
|
Name |
Lead bis(2-ethylhexanoate)
|
| Molecular Formula | C16H30O4Pb | |
| IUPAC Name* |
2-ethylhexanoate;lead(2+)
|
|
| SMILES |
CCCCC(CC)C(=O)[O-].CCCCC(CC)C(=O)[O-].[Pb+2]
|
|
| InChI |
InChI=1S/2C8H16O2.Pb/c2*1-3-5-6-7(4-2)8(9)10;/h2*7H,3-6H2,1-2H3,(H,9,10);/q;;+2/p-2
|
|
| InChIKey |
RUCPTXWJYHGABR-UHFFFAOYSA-L
|
|
| Synonyms |
Lead bis(2-ethylhexanoate); 301-08-6; Lead(II) 2-ethylhexanoate; 2-ethylhexanoate;lead(2+); Hexanoic acid, 2-ethyl-, lead(2+) salt (2:1); Hexanoic acid, 2-ethyl-, lead salt; Lead tallate 2-ethylhexanoate; EINECS 206-107-0; EINECS 269-090-9; MFCD00014003; SCHEMBL43687; SCHEMBL2423179; DTXSID50890498; 2-Ethylhexanoic acid lead(II) salt; Bis(2-ethylhexanoic acid)lead(II) salt; 68187-37-1; DB-047708; FT-0631914
|
|
| CAS | 301-08-6 | |
| PubChem CID | 160451 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 493.0 | ALogp: | -0.1 |
| HBD: | 0 | HBA: | 4 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 80.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 21 | QED Weighted: | 0.406 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.279 | MDCK Permeability: | 0.00003020 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.17 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.996 | Plasma Protein Binding (PPB): | 60.36% |
| Volume Distribution (VD): | 0.713 | Fu: | 39.91% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.063 | CYP1A2-substrate: | 0.48 |
| CYP2C19-inhibitor: | 0.028 | CYP2C19-substrate: | 0.883 |
| CYP2C9-inhibitor: | 0.103 | CYP2C9-substrate: | 0.925 |
| CYP2D6-inhibitor: | 0.005 | CYP2D6-substrate: | 0.143 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.085 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.138 | Half-life (T1/2): | 0.674 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.025 | Human Hepatotoxicity (H-HT): | 0.697 |
| Drug-inuced Liver Injury (DILI): | 0.037 | AMES Toxicity: | 0.024 |
| Rat Oral Acute Toxicity: | 0.059 | Maximum Recommended Daily Dose: | 0.03 |
| Skin Sensitization: | 0.737 | Carcinogencity: | 0.444 |
| Eye Corrosion: | 0.981 | Eye Irritation: | 0.988 |
| Respiratory Toxicity: | 0.704 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000628 | 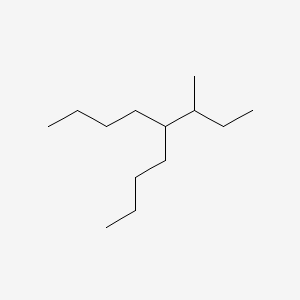 |
0.443 | D03LGY | 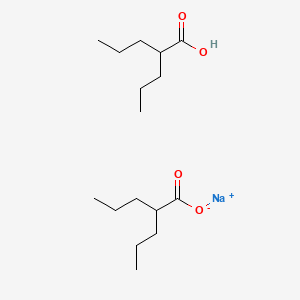 |
0.472 | ||
| ENC000213 | 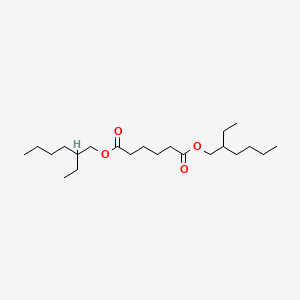 |
0.404 | D0X4FM | 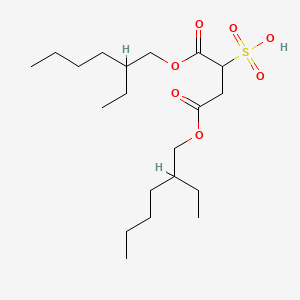 |
0.391 | ||
| ENC001132 | 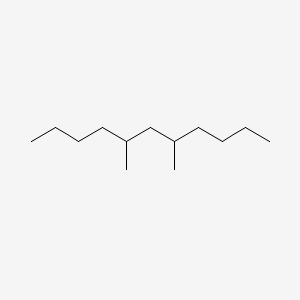 |
0.397 | D0Y3KG | 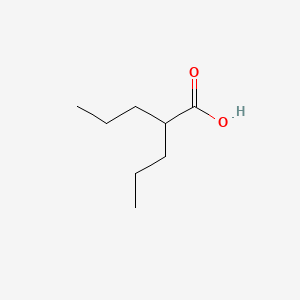 |
0.274 | ||
| ENC002444 | 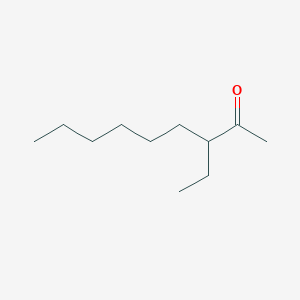 |
0.393 | D0AY9Q | 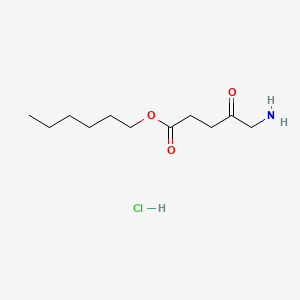 |
0.267 | ||
| ENC003061 | 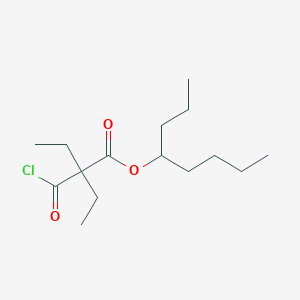 |
0.392 | D08EVN | 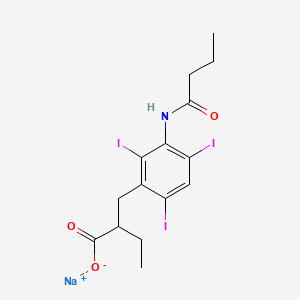 |
0.258 | ||
| ENC001041 |  |
0.391 | D0D9NY | 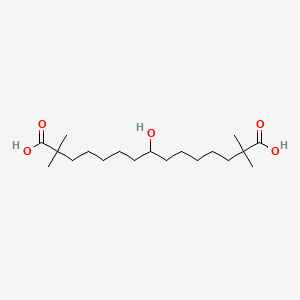 |
0.247 | ||
| ENC000290 |  |
0.379 | D00MLW |  |
0.245 | ||
| ENC001248 |  |
0.375 | D0ZI4H |  |
0.245 | ||
| ENC002794 | 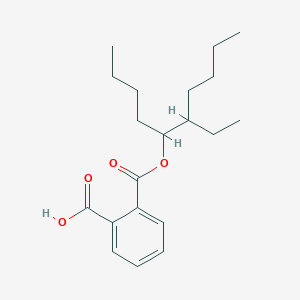 |
0.368 | D06ORU | 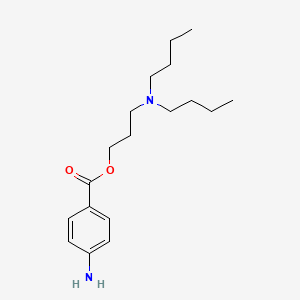 |
0.223 | ||
| ENC000833 | 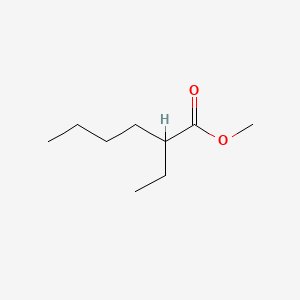 |
0.367 | D0N3NO | 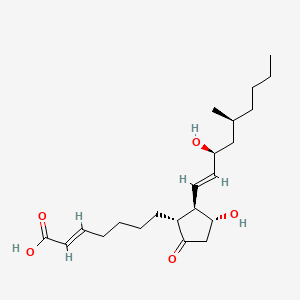 |
0.221 | ||