NPs Basic Information
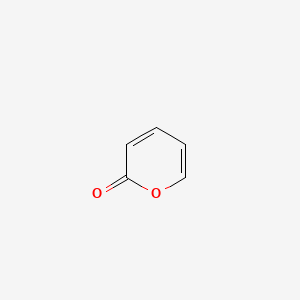
|
Name |
2H-Pyran-2-one
|
| Molecular Formula | C5H4O2 | |
| IUPAC Name* |
pyran-2-one
|
|
| SMILES |
C1=CC(=O)OC=C1
|
|
| InChI |
InChI=1S/C5H4O2/c6-5-3-1-2-4-7-5/h1-4H
|
|
| InChIKey |
ZPSJGADGUYYRKE-UHFFFAOYSA-N
|
|
| Synonyms |
2H-Pyran-2-one; alpha-Pyrone; 504-31-4; pyran-2-one; Pyranone; 2-Pyranone; 2-Pyrone; Coumalin; Pyrone; a-pyrone; .alpha.-Pyrone; 2-oxo-2H-pyran; CHEMBL1934663; CHEBI:37965; 8WW45I202V; oxidopyrylium; UNII-8WW45I202V; EINECS 207-990-5; 2H-Pyrane-2-one; 1,2-Pyrone; 2H-Pyran, 2-oxo-; 2H-Pyran-2-one, 90%; DTXSID50198441; ZINC1846601; BDBM50360796; GEO-04164; MFCD00006639; CS-W013734; AS-56552; DB-051773; FT-0622198; P1183; 2H-Pyran-2-one, technical, >=90% (GC); 5-hydroxy-2,4-pentadienoic acid delta-lactone; F16388; 5-hydroxy-2,4-pentadienoic acid delta -lactone; A828125; Q209475; 2,4-Pentadienoic acid, 5-hydroxy-, .delta.-lactone
|
|
| CAS | 504-31-4 | |
| PubChem CID | 68154 | |
| ChEMBL ID | CHEMBL1934663 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 96.08 | ALogp: | 0.8 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 1 |
| Heavy Atoms: | 7 | QED Weighted: | 0.483 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.308 | MDCK Permeability: | 0.00003240 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.37 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.655 |
| 30% Bioavailability (F30%): | 0.261 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.36 | Plasma Protein Binding (PPB): | 72.93% |
| Volume Distribution (VD): | 1.079 | Fu: | 40.30% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.741 | CYP1A2-substrate: | 0.511 |
| CYP2C19-inhibitor: | 0.135 | CYP2C19-substrate: | 0.09 |
| CYP2C9-inhibitor: | 0.016 | CYP2C9-substrate: | 0.113 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.408 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.227 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.055 | Half-life (T1/2): | 0.822 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.141 | Human Hepatotoxicity (H-HT): | 0.081 |
| Drug-inuced Liver Injury (DILI): | 0.214 | AMES Toxicity: | 0.068 |
| Rat Oral Acute Toxicity: | 0.867 | Maximum Recommended Daily Dose: | 0.02 |
| Skin Sensitization: | 0.209 | Carcinogencity: | 0.874 |
| Eye Corrosion: | 0.989 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.883 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000190 | 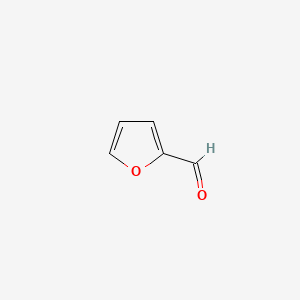 |
0.379 | D0X9RY | 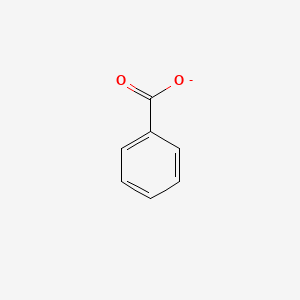 |
0.286 | ||
| ENC000675 | 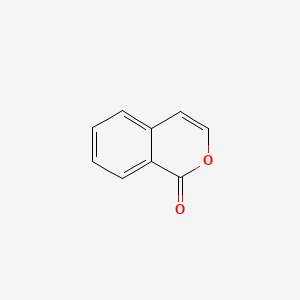 |
0.368 | D07HBX |  |
0.270 | ||
| ENC000480 | 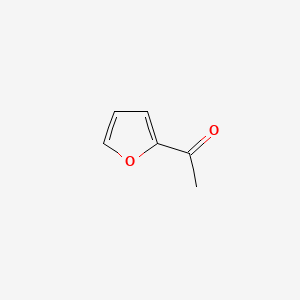 |
0.355 | D0D5GG | 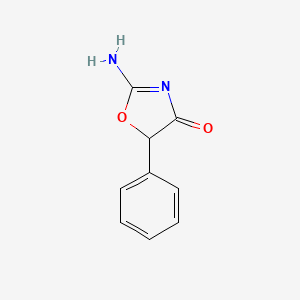 |
0.267 | ||
| ENC000162 | 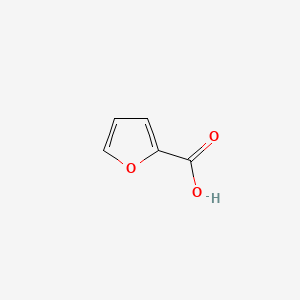 |
0.355 | D05OIS | 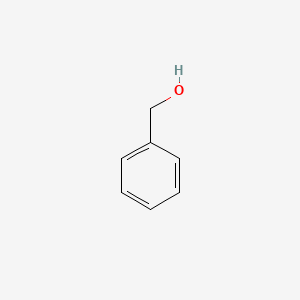 |
0.265 | ||
| ENC000025 | 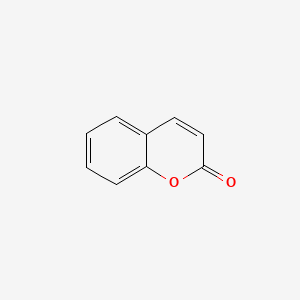 |
0.333 | D03GET | 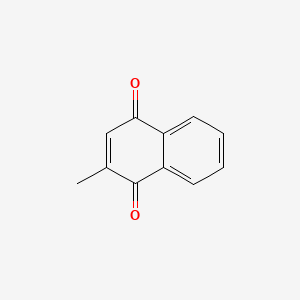 |
0.244 | ||
| ENC000189 |  |
0.333 | D01ZJK | 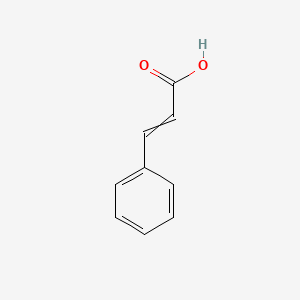 |
0.244 | ||
| ENC000243 | 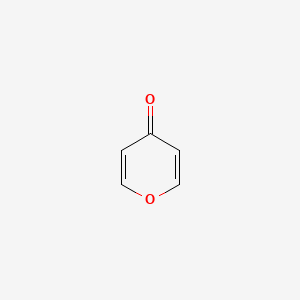 |
0.333 | D06DLI | 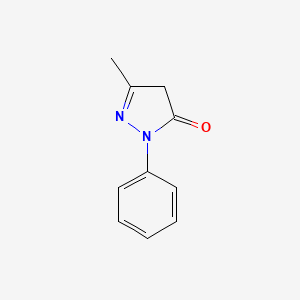 |
0.239 | ||
| ENC000681 | 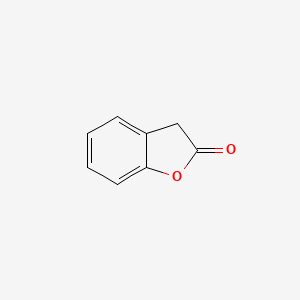 |
0.324 | D0R1CR | 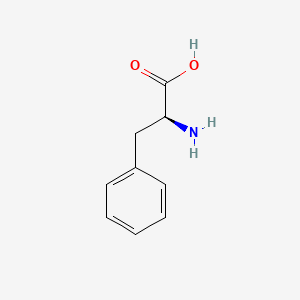 |
0.233 | ||
| ENC001133 | 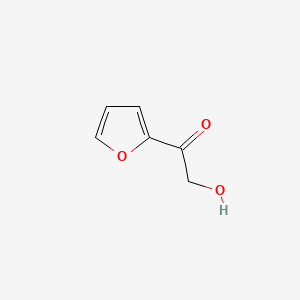 |
0.324 | D05BMG | 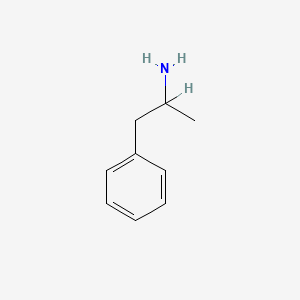 |
0.231 | ||
| ENC002806 | 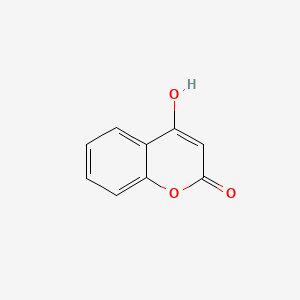 |
0.317 | D0T3LF | 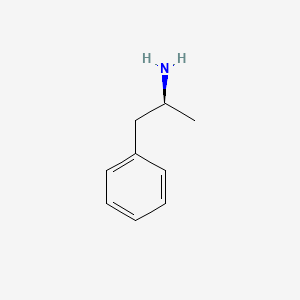 |
0.231 | ||