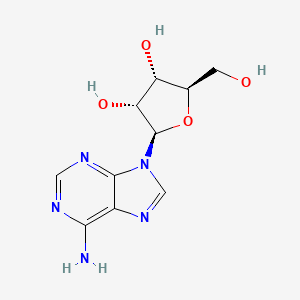
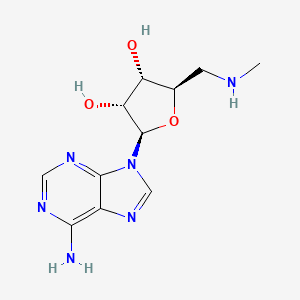
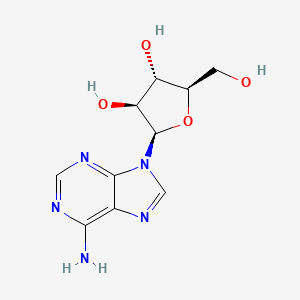
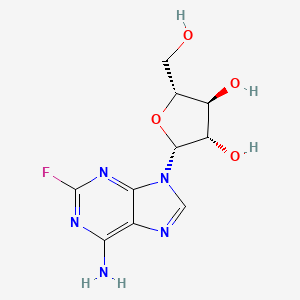
| InChI |
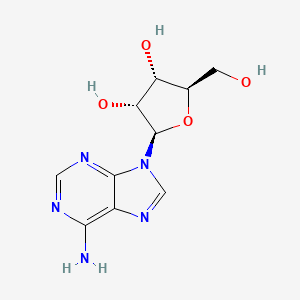
InChI=1S/C10H13N5O4/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(18)6(17)4(1-16)19-10/h2-4,6-7,10,16-18H,1H2,(H2,11,12,13)/t4-,6-,7-,10-/m1/s1
|
| Synonyms |
adenosine; 58-61-7; Adenocard; Adenoscan; Adenine riboside; beta-D-Adenosine; Nucleocardyl; Adenosin; Boniton; Sandesin; Myocol; Adenine nucleoside; Adenocor; beta-Adenosine; 9-beta-D-Ribofuranosyladenine; 9-beta-D-Ribofuranosidoadenine; 9-beta-D-Ribofuranosyl-9H-purin-6-amine; Adenosin [German]; USAF CB-10; 9beta-D-Ribofuranosyladenine; 6-Amino-9-beta-D-ribofuranosyl-9H-purine; Ade-Rib; Caswell No. 010B; (2R,3R,4S,5R)-2-(6-Aminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol; SR 96225; Adenosine [USAN:BAN]; (2R,3R,4S,5R)-2-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol; beta-D-Ribofuranoside, adenine-9; 6-Amino-9beta-D-ribofuranosyl-9H-purine; (2R,3R,4S,5R)-2-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol; CHEBI:16335; Quinquefolan B; 3H-adenosine; NSC 7652; Dehydran 240; SR-96225; 9-.beta.-d-Ribofuranosyladenine; Adenosine (Adenocard); AI3-52413; 9H-Purin-6-amine, 9beta-D-ribofuranosyl-; CHEMBL477; D-Adenosine; beta-D-Ribofuranose, 1-(6-amino-9H-purin-9-yl)-1-deoxy-; adenine-D-ribose; K72T3FS567; NSC-7652; CCRIS 2557; NCGC00023673-05; Pallacor; 41547-82-4; MFCD00005752; DSSTox_CID_2558; DSSTox_RID_76628; DSSTox_GSID_22558; 109767-06-8; 133248-01-8; .beta.-D-Adenosine; CAS-58-61-7; Adenocard (TN); Adenoscan (TN); SMR000058216; MEDR-640; Adenosine (JAN/USP); SR-05000001981; EINECS 200-389-9; Adenine-beta-D-arabinofuranoside; NSC 627048; NSC7652; Adenogesic; Adenosine [USAN:USP:BAN]; Adenin riboside; UNII-K72T3FS567; .beta.-D-Ribofuranoside, adenine-9; 9-.alpha.-D-Arabinofuranosyladenine; NSC627048; b-D-Adenosine; HSDB 7774; SUN-Y4001; N6-Methylado; 1dgm; 1odi; 2fqy; 2ydo; 3axz; 4cki; 4ckj; Adenosine,(S); beta-delta-Adenosine; [U-14C]adenosine; 6-Amino-9.beta.-D-ribofuranosyl-9H-purine; 2gl0; 3ay0; 4pd9; Adenosine, >=99%; ADENOSINE [JAN]; ADENOSINE [MI]; 6-Amino-9-.beta.-ribofuranosyl-9H-purine; ADENOSINE [HSDB]; ADENOSINE [INCI]; ADENOSINE [USAN]; Spectrum2_001257; Spectrum3_000288; ADENOSINE [VANDF]; cid_191; SCHEMBL731; ADENOSINE [MART.]; bmse000061; bmse000996; Epitope ID:140947; ADENOSINE [USP-RS]; ADENOSINE [WHO-DD]; 4-Aminopyrazolo[3,4-d]pyrimidine ribonucleoside; 9-ss-D-Ribofuranosyladenine; BSPBio_001796; .beta.-D-Ribofuranose, 1-(6-amino-9H-purin-9-yl)-1-deoxy-; cid_60961; MLS000069638; MLS002153227; MLS006010946; SPECTRUM1500107; Adenine-9-ss-D-ribofuranoside; REGID_for_CID_60961; SPBio_001194; adenine-9beta-D-Ribofuranoside; GTPL2844; 9beta-delta-Ribofuranosyladenine; ADENOSINE [EP IMPURITY]; ADENOSINE [ORANGE BOOK]; ADENOSINE [EP MONOGRAPH]; DTXSID1022558; BDBM14487; KBio3_001296; 9-beta-delta-Ribofuranosyladenine; ADENOSINE [USP MONOGRAPH]; EA6C60C2-6AFB-4264-A2F0-541373DB950E; 9-.beta.-D-Ribofuranosidoadenine; 9-beta-delta-Ribofuranosidoadenine; adenine-9beta-delta-Ribofuranoside; Bio1_000437; Bio1_000926; Bio1_001415; HMS1920A13; HMS2091G13; HMS2235E24; HMS3884O04; Pharmakon1600-01500107; ACT02616; ALBB-032827; AMY30083; ZINC2169830; 9-beta-delta-Arabinofuranosyladenine; Tox21_110891; AC7861; CCG-38824; NSC755857; s1647; AKOS015888594; Tox21_110891_1; AC-8229; AM83931; DB00640; NSC-755857; SDCCGMLS-0003108.P003; 9-?-D-Ribofuranosyl-9H-purin-6-amine; 9beta-D-ribofuranosyl-9H-Purin-6-amine; NCGC00023673-03; NCGC00023673-04; NCGC00023673-06; NCGC00023673-07; NCGC00023673-10; NCGC00023673-20; NCGC00178869-03; AC-27494; AS-12664; Adenosine, Vetec(TM) reagent grade, 98%; SBI-0206673.P002; 9beta-delta-ribofuranosyl-9H-Purin-6-amine; DB-022408; 6-Amino-9beta-delta-ribofuranosyl-9H-purine; 9-.beta.-D-Ribofuranosyl-9H-purin-6-amine; 9-beta-delta-Ribofuranosyl-9H-purin-6-amine; A0152; 9H-Purin-6-amine, 9-.beta.-d-ribofuranosyl-; C00212; D00045; EN300-100931; AB00384349-11; AB00384349_13; AB00384349_14; Adenosine, BioReagent, suitable for cell culture; Q190012; 6-AMINO-9-.BETA.-D-RIBOFURANOSYL-9H-PURINE; SR-05000001981-1; SR-05000001981-2; Z1341543331; 1-(6-amino-9H-purin-9-yl)-1-deoxy-beta-D-Ribofuranose; Adenosine, European Pharmacopoeia (EP) Reference Standard; Formycin A, from Streptomyces kaniharaensis, >=98% (HPLC); 1-(6-amino-9H-purin-9-yl)-1-deoxy-beta-delta-Ribofuranose; Adenosine, United States Pharmacopeia (USP) Reference Standard; Adenosine, Pharmaceutical Secondary Standard; Certified Reference Material; (2R,3R,4S,5R)-2-(6-aminopurin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol; 142796-17-6
|