NPs Basic Information
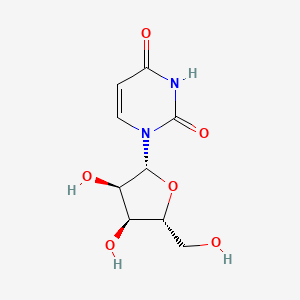
|
Name |
Uridine
|
| Molecular Formula | C9H12N2O6 | |
| IUPAC Name* |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2,4-dione
|
|
| SMILES |
C1=CN(C(=O)NC1=O)[C@H]2[C@@H]([C@@H]([C@H](O2)CO)O)O
|
|
| InChI |
InChI=1S/C9H12N2O6/c12-3-4-6(14)7(15)8(17-4)11-2-1-5(13)10-9(11)16/h1-2,4,6-8,12,14-15H,3H2,(H,10,13,16)/t4-,6-,7-,8-/m1/s1
|
|
| InChIKey |
DRTQHJPVMGBUCF-XVFCMESISA-N
|
|
| Synonyms |
uridine; 58-96-8; Uridin; Uracil riboside; 1-beta-D-Ribofuranosyluracil; Uracil, 1-beta-D-ribofuranosyl-; beta-Uridine; d-uridine; b-Uridine; NSC 20256; 1-.beta.-D-Ribofuranosyluracil; CHEBI:16704; Urd; AI3-52690; 1-((2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)pyrimidine-2,4(1H,3H)-dione; WHI7HQ7H85; MLS000069625; NSC-20256; SMR000058222; MFCD00006526; 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2,4-dione; 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,3,4-tetrahydropyrimidine-2,4-dione; 1-beta-D-ribofuranosylpyrimidine-2,4(1H,3H)-dione; 1-beta-D-Ribofuranosylpyrimidine-2,4(1H,3H)-dione (Uridine); 1-((2R,3R,4S,5R)-tetrahydro-3,4-dihydroxy-5-(hydroxymethyl)furan-2-yl)pyrimidine-2,4(1H,3H)-dione; 3083-77-0; EINECS 200-407-5; UNII-WHI7HQ7H85; araU; NSC20256; d-Ribosyl uracil; .beta.-Uridine; Uridine,(S); 1af2; 4jx9; 4pd6; Opera_ID_118; URIDINE [INCI]; Uridine, >=99%; URIDINE [MI]; URIDINE [MART.]; 1-b-D-Ribofuranosyluracil; URIDINE [USP-RS]; URIDINE [WHO-DD]; 1-A-D-Ribofuranosyluracil; bmse000158; bmse000816; bmse000864; Epitope ID:149164; 1-.beta.-D-Ribofuranosyl-2,4(1H,3H)-pyrimidinedione; beta-D-ribofuranosyl-uridine; SCHEMBL20667; Uracil-1-A-D-ribofuranoside; rg2417; Uridine, BioUltra, >=99%; URIDINE [USP IMPURITY]; CHEMBL100259; GTPL4566; 1-beta-delta-Ribofuranosyluracil; DTXSID40891555; Uracil-1-.beta.-d-ribofuranoside; HMS2230P13; HMS3884M20; HY-B1449; ZINC2583633; BDBM50088517; s2029; AKOS015896922; AM83934; CCG-214447; CS-5153; DB02745; SMP1_000029; NCGC00017312-02; NCGC00017312-04; NCGC00017312-05; NCGC00142368-01; 1-[(4S,2R,3R,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,3-dihydropyrimi dine-2,4-dione; DS-14345; SRI-10895_12; Uridine, Vetec(TM) reagent grade, 99%; ADENOSINE IMPURITY F [EP IMPURITY]; DB-030519; U0020; C00299; EN300-221746; 1-b-D-Ribofuranosyl-2,4(1H,3H)-pyrimidinedione; b-D-Ribofuranoside 2,4(1H,3H)-pyrimidinedione-1; Q422573; BRD-K13050303-001-18-1; Uridine, powder, BioReagent, suitable for cell culture; .beta.-D-Ribofuranoside, 2,4(1H,3H)-pyrimidinedione-1; 1-beta-delta-Ribofuranosyl-2,4(1H,3H)-pyrimidinedione; 6B6FA3F8-70A2-44EA-B99C-D35D0A9237AA; beta-delta-Ribofuranoside 2,4(1H,3H)-pyrimidinedione-1; Z1741976658; 1-.BETA.-D-RIBOFURANOSYLPYRIMIDINE-2,4(1H,3H)-DIONE; 1-beta-D-Ribofuranosyluracil, Uracil-1-beta-D-ribofuranoside; Uridine, United States Pharmacopeia (USP) Reference Standard; 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]pyrimidine-2,4-dione
|
|
| CAS | 58-96-8 | |
| PubChem CID | 6029 | |
| ChEMBL ID | CHEMBL100259 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 244.2 | ALogp: | -2.0 |
| HBD: | 4 | HBA: | 6 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 119.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 17 | QED Weighted: | 0.464 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -6.077 | MDCK Permeability: | 0.00023342 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.01 |
| Human Intestinal Absorption (HIA): | 0.856 | 20% Bioavailability (F20%): | 0.839 |
| 30% Bioavailability (F30%): | 0.982 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.41 | Plasma Protein Binding (PPB): | 13.82% |
| Volume Distribution (VD): | 0.385 | Fu: | 79.15% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.011 | CYP1A2-substrate: | 0.083 |
| CYP2C19-inhibitor: | 0.024 | CYP2C19-substrate: | 0.047 |
| CYP2C9-inhibitor: | 0.001 | CYP2C9-substrate: | 0.092 |
| CYP2D6-inhibitor: | 0.015 | CYP2D6-substrate: | 0.093 |
| CYP3A4-inhibitor: | 0.004 | CYP3A4-substrate: | 0.024 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.413 | Half-life (T1/2): | 0.847 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.756 |
| Drug-inuced Liver Injury (DILI): | 0.986 | AMES Toxicity: | 0.096 |
| Rat Oral Acute Toxicity: | 0.004 | Maximum Recommended Daily Dose: | 0.005 |
| Skin Sensitization: | 0.043 | Carcinogencity: | 0.034 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.015 |
| Respiratory Toxicity: | 0.012 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005638 | 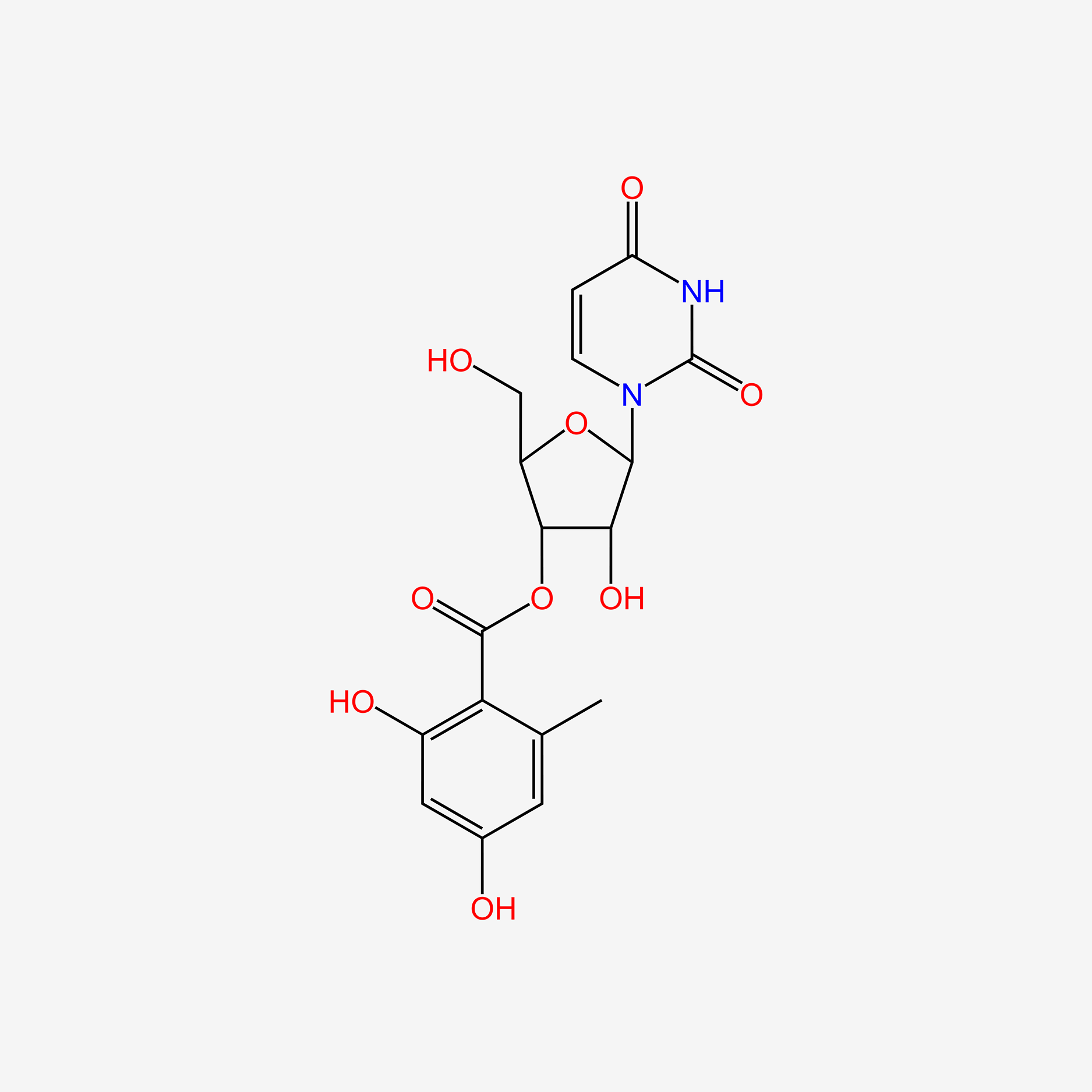 |
0.506 | D0Y7DP | 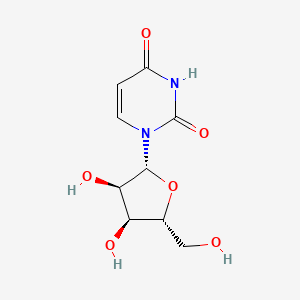 |
1.000 | ||
| ENC005639 | 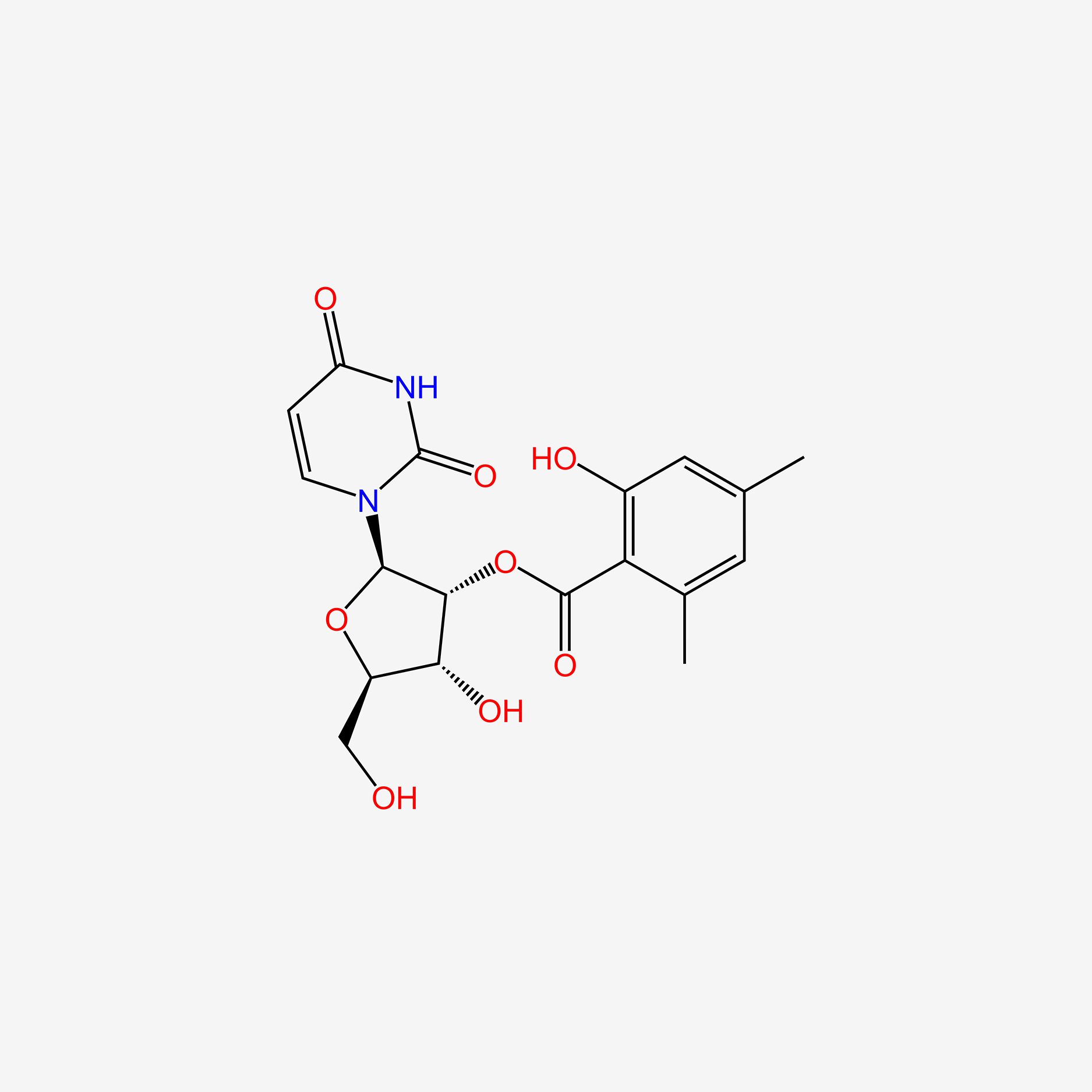 |
0.506 | D07XSN |  |
0.614 | ||
| ENC002576 | 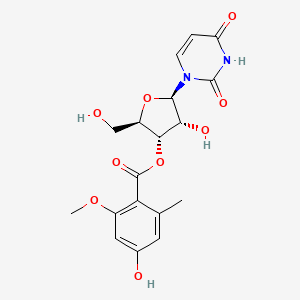 |
0.488 | D03TGJ | 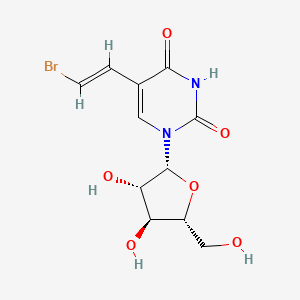 |
0.587 | ||
| ENC000635 | 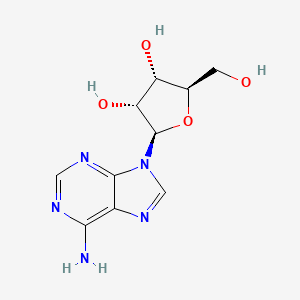 |
0.435 | D09FAZ | 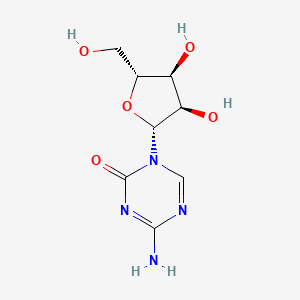 |
0.508 | ||
| ENC000120 | 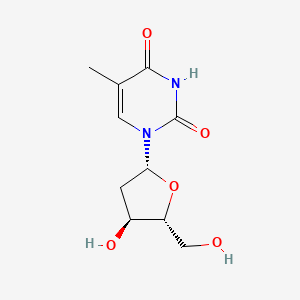 |
0.394 | D0G5AG | 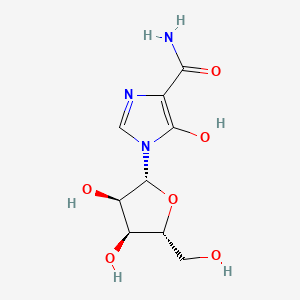 |
0.469 | ||
| ENC000661 | 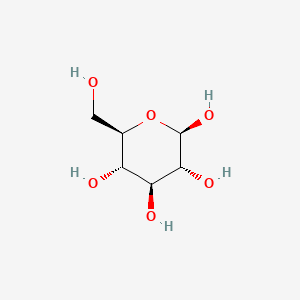 |
0.351 | D03KXY | 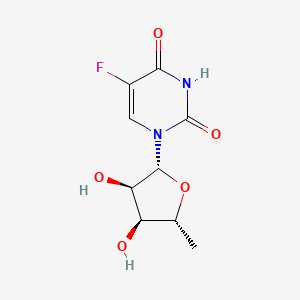 |
0.444 | ||
| ENC005772 | 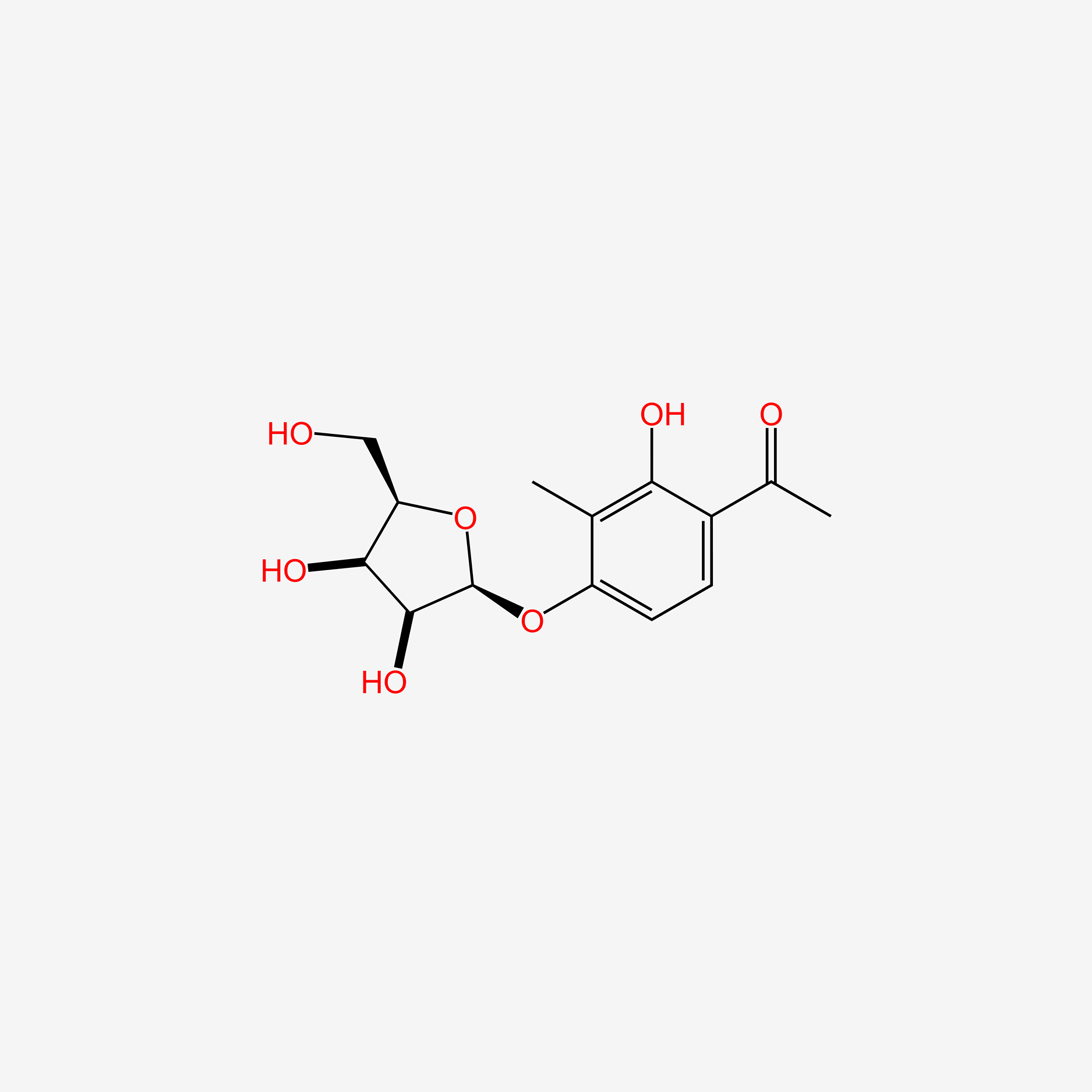 |
0.342 | D06IAR | 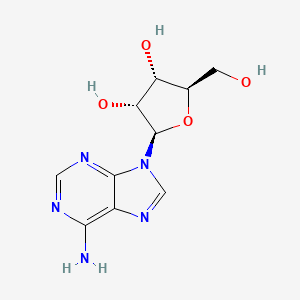 |
0.435 | ||
| ENC004076 | 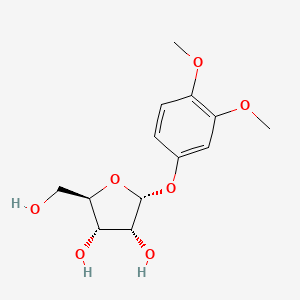 |
0.329 | D0NI0C | 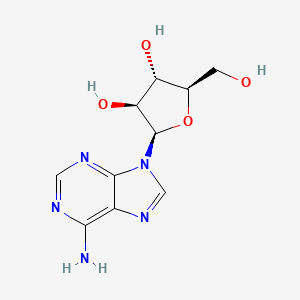 |
0.435 | ||
| ENC001067 | 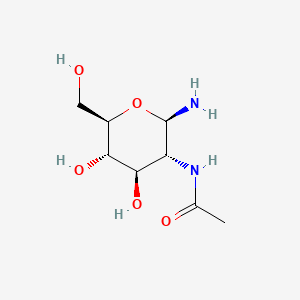 |
0.328 | D0H3WI | 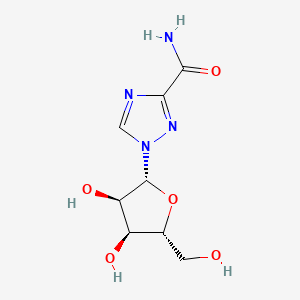 |
0.415 | ||
| ENC002667 | 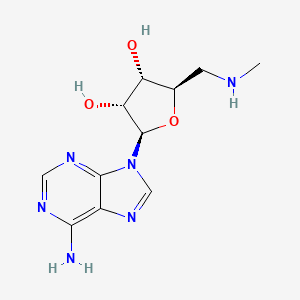 |
0.325 | D0F2XQ | 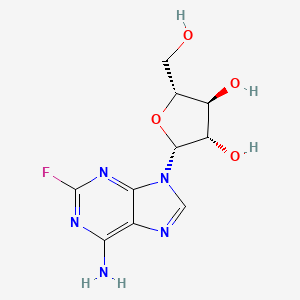 |
0.403 | ||