NPs Basic Information
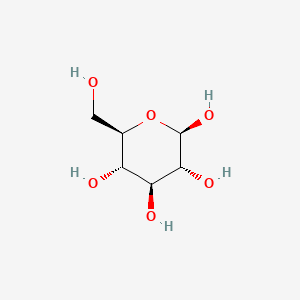
|
Name |
(2R,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol
|
| Molecular Formula | C6H12O6 | |
| IUPAC Name* |
(2R,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol
|
|
| SMILES |
C([C@@H]1[C@H]([C@@H]([C@H]([C@@H](O1)O)O)O)O)O
|
|
| InChI |
InChI=1S/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3-,4+,5-,6-/m1/s1
|
|
| InChIKey |
WQZGKKKJIJFFOK-VFUOTHLCSA-N
|
|
| Synonyms |
beta-D-glucose; beta-D-glucopyranose; 492-61-5; glucoside; Curdlan; beta-Dextrose; beta-glucose; b-d-glucose; 28905-12-6; .beta.-D-Glucopyranose; 54724-00-4; .beta.-D-Glucose; (2R,3R,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol; (2R,3R,4S,5S,6R)-6-(Hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetraol; .beta.-d-Glucose, anhydrous; Grape sugar; Corn sugar; CHEMBL1614854; CHEBI:15903; J4R00M814D; (+)-Glucose; 4-Morpholineacetic acid, a-Methylene-, Methyl ester; 128009-02-9; 133947-06-5; Glucodin; Goldsugar; Meritose; 136760-05-9; Vadex; Clintose L; ZYMOSAN; CPC hydrate; Roferose ST; Clearsweet 95; Glucosides; Staleydex 95M; Staleydex 111; Oxidase, glucose; Cerelose 2001; .beta.-D-ribo-Hexopyranose, 1,6-anhydro-3-deoxy-2-O-phenyl-4-O-(phenylmethyl)-; Tabfine 097(HS); BGC; 9001-37-0; Glucose, (beta-D)-Isomer; Callose; UNII-J4R00M814D; beta-D-Glucopyranose, anhydrous; beta -d-glucose; beta-D-Glc; EINECS 207-756-2; 9010-72-4; beta-d-(+)-glucose; 1,3-beta-D-Glucan; beta-(1,3)-glucan; I(2)-D-Glucopyranose; Beta-D-glucose anhydrous; Beta-d-glucose, anhydrous; 6-(hydroxymethyl)tetrahydro-2H-pyran-2,3,4,5-tetrol; GLUCOSE, BETA-D-; (1,2-beta-D-glucosyl)n; (1->2)-beta-D-glucan; (1->3)-beta-D-glucan; (1->4)-beta-D-glucan; (1->6)-beta-D-glucan; GLUCOSE, .BETA.-D; SCHEMBL25601; (1->2)-beta-D-glucopyranan; (1->3)-beta-D-glucopyranan; (1->4)-beta-D-glucopyranan; (1->6)-beta-D-glucopyranan; CHEBI:18246; CHEBI:27380; CHEBI:27517; CHEBI:37671; DTXSID70883403; Pharmakon1600-01300015; .BETA.-D-GLUCOSE ANHYDROUS; ZINC3833800; beta-D-glucose; D-glucose; glucose; BDBM50240803; MFCD00063989; NSC759603; AKOS016010209; DB02379; NSC-759603; YC46078; NCGC00263446-02; Beta-D-glucose(contains alpha-D-glucose); BS-22220; G0047; C00221; D90709; AE-562/43459286; BETA-D-GLUCOSE (CONTAINS ALPHA-D-GLUCOSE); W-202206; Q23905968; WURCS=2.0/1,1,0/[a2122h-1b_1-5]/1/; 50986-29-3
|
|
| CAS | 492-61-5 | |
| PubChem CID | 64689 | |
| ChEMBL ID | CHEMBL1614854 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 180.16 | ALogp: | -2.6 |
| HBD: | 5 | HBA: | 6 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 110.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 12 | QED Weighted: | 0.299 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.39 | MDCK Permeability: | 0.00032048 |
| Pgp-inhibitor: | 0.004 | Pgp-substrate: | 0.109 |
| Human Intestinal Absorption (HIA): | 0.836 | 20% Bioavailability (F20%): | 0.036 |
| 30% Bioavailability (F30%): | 0.984 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.301 | Plasma Protein Binding (PPB): | 13.65% |
| Volume Distribution (VD): | 0.411 | Fu: | 79.72% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.024 | CYP1A2-substrate: | 0.048 |
| CYP2C19-inhibitor: | 0.017 | CYP2C19-substrate: | 0.216 |
| CYP2C9-inhibitor: | 0.001 | CYP2C9-substrate: | 0.129 |
| CYP2D6-inhibitor: | 0.005 | CYP2D6-substrate: | 0.12 |
| CYP3A4-inhibitor: | 0.005 | CYP3A4-substrate: | 0.017 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.602 | Half-life (T1/2): | 0.821 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.061 | Human Hepatotoxicity (H-HT): | 0.049 |
| Drug-inuced Liver Injury (DILI): | 0.044 | AMES Toxicity: | 0.457 |
| Rat Oral Acute Toxicity: | 0.038 | Maximum Recommended Daily Dose: | 0.002 |
| Skin Sensitization: | 0.237 | Carcinogencity: | 0.03 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.733 |
| Respiratory Toxicity: | 0.028 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003055 |  |
0.591 | D0H3KI |  |
1.000 | ||
| ENC001062 |  |
0.581 | D07NSU | 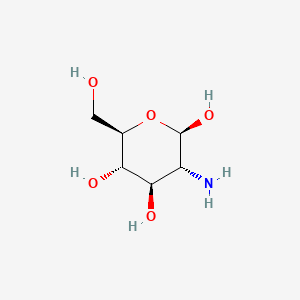 |
0.676 | ||
| ENC003068 | 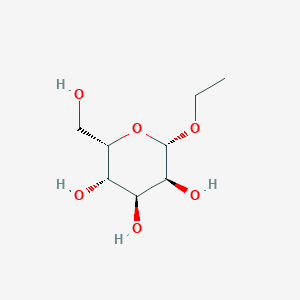 |
0.581 | D0H2RI | 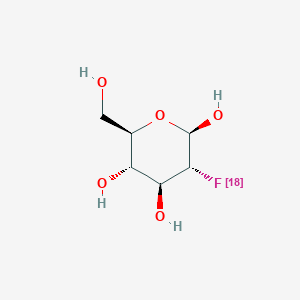 |
0.676 | ||
| ENC000851 | 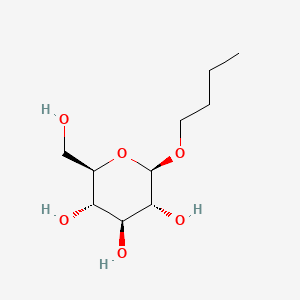 |
0.510 | D05ZYM | 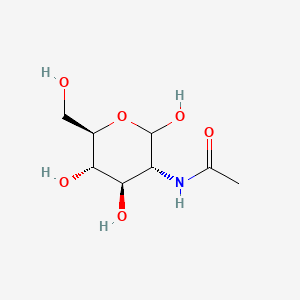 |
0.556 | ||
| ENC003177 | 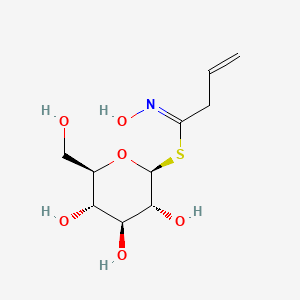 |
0.491 | D0Z4EI | 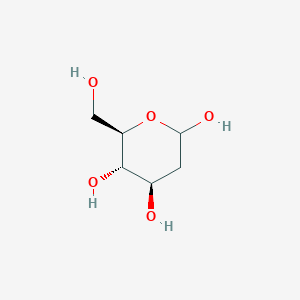 |
0.500 | ||
| ENC001067 | 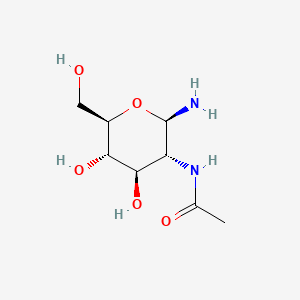 |
0.429 | D0I8RR |  |
0.472 | ||
| ENC004291 |  |
0.426 | D06BQU |  |
0.433 | ||
| ENC003363 |  |
0.404 | D07HZY |  |
0.425 | ||
| ENC005608 |  |
0.403 | D0T5BC |  |
0.409 | ||
| ENC001625 |  |
0.388 | D0MU9L | 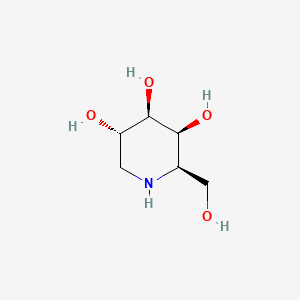 |
0.364 | ||