NPs Basic Information
|
Name |
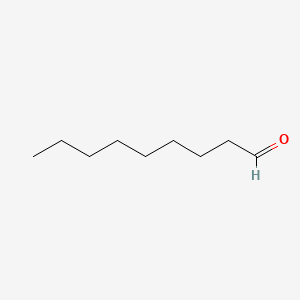
Nonanal
|
| Molecular Formula | C9H18O | |
| IUPAC Name* |
nonanal
|
|
| SMILES |
CCCCCCCCC=O
|
|
| InChI |
InChI=1S/C9H18O/c1-2-3-4-5-6-7-8-9-10/h9H,2-8H2,1H3
|
|
| InChIKey |
GYHFUZHODSMOHU-UHFFFAOYSA-N
|
|
| Synonyms |
NONANAL; 124-19-6; Pelargonaldehyde; 1-Nonanal; Nonanaldehyde; Nonyl aldehyde; Nonaldehyde; n-Nonaldehyde; Pelargonic aldehyde; Nonylic aldehyde; n-Nonanal; Nonylaldehyde; Aldehyde C-9; 1-Nonaldehyde; Nonanoic aldehyde; 1-Nonyl aldehyde; C-9 aldehyde; n-Nonylaldehyde; NONYL ALDEHYDE,N-; FEMA No. 2782; NCI-C61018; NSC 5518; MFCD00007030; 2L2WBY9K6T; CHEBI:84268; NSC5518; NSC-5518; Nonoic aldehyde; Aldehyde C9; CCRIS 664; HSDB 7229; n-NONYL ALDEHYDE; EINECS 204-688-5; UNII-2L2WBY9K6T; BRN 1236701; non-aldehyde; aldehyde c; AI3-04859; n-Nonan-1-al; C9-11 Aldehydes; C9-11-Aldehydes; Nonyl aldehyde, n-; Nonanal, 95%; C9 ALDEHYDE; NONANAL [HSDB]; NONANAL [FCC]; DSSTox_CID_1639; N-NONANAL [FHFI]; EC 204-688-5; WLN: VH8; Nonanal, analytical standard; DSSTox_RID_76253; DSSTox_GSID_21639; SCHEMBL22860; Nonanal, >=95%, FCC; 4-01-00-03352 (Beilstein Handbook Reference); QSPL 015; SCHEMBL8876408; CHEMBL2228376; DTXSID9021639; Nonanal, natural, >=98%, FG; AMY15728; HY-N8016; ZINC1686990; EINECS 278-296-8; Tox21_303603; LMFA06000040; AKOS009158987; FS-3913; NCGC00257442-01; BP-31179; CAS-124-19-6; SY016777; DB-041769; CS-0138979; FT-0631724; N0296; Aldehyde C9, Nonyl aldehyde, Pelargonaldehyde; EN300-135251; Q419668; J-005053
|
|
| CAS | 124-19-6 | |
| PubChem CID | 31289 | |
| ChEMBL ID | CHEMBL2228376 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 142.24 | ALogp: | 3.3 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.391 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.495 | MDCK Permeability: | 0.00001970 |
| Pgp-inhibitor: | 0.026 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.99 |
| 30% Bioavailability (F30%): | 0.991 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.994 | Plasma Protein Binding (PPB): | 47.28% |
| Volume Distribution (VD): | 1.857 | Fu: | 41.43% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.809 | CYP1A2-substrate: | 0.608 |
| CYP2C19-inhibitor: | 0.249 | CYP2C19-substrate: | 0.356 |
| CYP2C9-inhibitor: | 0.175 | CYP2C9-substrate: | 0.842 |
| CYP2D6-inhibitor: | 0.05 | CYP2D6-substrate: | 0.254 |
| CYP3A4-inhibitor: | 0.058 | CYP3A4-substrate: | 0.103 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.601 | Half-life (T1/2): | 0.526 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.181 | Human Hepatotoxicity (H-HT): | 0.021 |
| Drug-inuced Liver Injury (DILI): | 0.047 | AMES Toxicity: | 0.175 |
| Rat Oral Acute Toxicity: | 0.042 | Maximum Recommended Daily Dose: | 0.026 |
| Skin Sensitization: | 0.964 | Carcinogencity: | 0.43 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.983 |
| Respiratory Toxicity: | 0.963 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000267 |  |
0.903 | D0Z5BC |  |
0.422 | ||
| ENC000032 |  |
0.893 | D05ATI |  |
0.389 | ||
| ENC000275 |  |
0.824 | D0Z5SM |  |
0.344 | ||
| ENC000277 |  |
0.757 | D0O1PH |  |
0.343 | ||
| ENC000460 |  |
0.697 | D0E4WR |  |
0.340 | ||
| ENC001601 |  |
0.676 | D0O1TC | 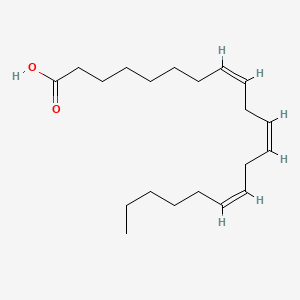 |
0.338 | ||
| ENC000607 | 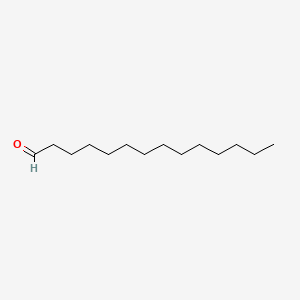 |
0.651 | D0Y8DP | 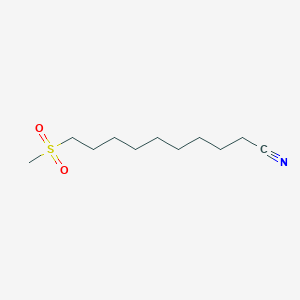 |
0.327 | ||
| ENC000268 | 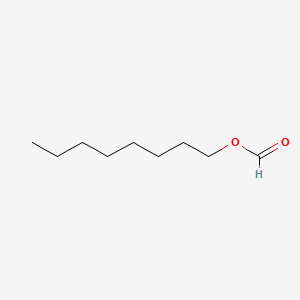 |
0.639 | D0AY9Q |  |
0.321 | ||
| ENC000455 |  |
0.639 | D0XN8C |  |
0.318 | ||
| ENC001684 |  |
0.639 | D03ZJE | 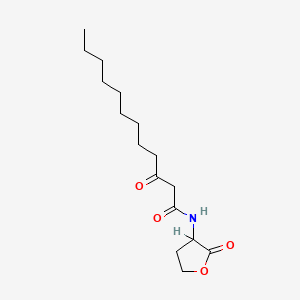 |
0.318 | ||