NPs Basic Information
|
Name |
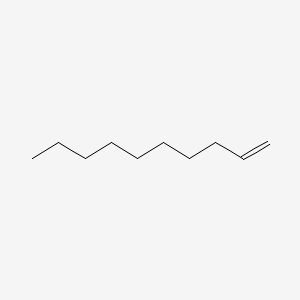
1-Decene
|
| Molecular Formula | C10H20 | |
| IUPAC Name* |
dec-1-ene
|
|
| SMILES |
CCCCCCCCC=C
|
|
| InChI |
InChI=1S/C10H20/c1-3-5-7-9-10-8-6-4-2/h3H,1,4-10H2,2H3
|
|
| InChIKey |
AFFLGGQVNFXPEV-UHFFFAOYSA-N
|
|
| Synonyms |
1-DECENE; Dec-1-ene; 872-05-9; Decylene; n-1-Decene; 1-n-Decene; alpha-Decene; n-Decylene; Gulftene 10; decene-1; 68037-01-4; .alpha.-Decene; NSC 62122; MFCD00009577; CHEBI:87315; 7O4U4C718P; NSC-62122; 25189-70-2; 37309-58-3; Dialene 10; Decene, n-; CCRIS 5718; HSDB 1073; EINECS 212-819-2; UNII-U333RI6EB7; UNII-7O4U4C718P; Linealene 10; Neodene 10; AHec-1-ene; DECENE [INCI]; 1-Decene, 94%; DSSTox_CID_7329; 1-DECENE [HSDB]; EC 212-819-2; EC 500-183-1; DECENE, 1-; DSSTox_RID_78410; UNII-4U179ML4TJ; UNII-CQ4IKK1766; Alkenes, C10-16 alpha-; DSSTox_GSID_27329; 1-Decene, analytical standard; UNII-75Y1X69O7I; 4U179ML4TJ; CQ4IKK1766; U333RI6EB7; CHEMBL3187990; DTXSID8027329; 1-DECENE MFC10 H20; 75Y1X69O7I; UNII-4YI0729529; NSC62122; ZINC1691021; EINECS 272-492-7; Tox21_200195; LMFA11000311; 1-Decene, >=97.0% (GC); AKOS015902910; 1-Decene, purum, >=95.0% (GC); NCGC00248557-01; NCGC00257749-01; 1-C10H20; CAS-872-05-9; DB-056981; 1-DECENE [STANDARD MATERIAL FOR GC]; 4YI0729529; D0028; FT-0607692; S0340; EN300-384588; T73018; Q151410; W-107244; F8881-0779
|
|
| CAS | 872-05-9 | |
| PubChem CID | 13381 | |
| ChEMBL ID | CHEMBL3187990 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 140.27 | ALogp: | 5.7 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.359 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.436 | MDCK Permeability: | 0.00001720 |
| Pgp-inhibitor: | 0.009 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.315 |
| 30% Bioavailability (F30%): | 0.617 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.991 | Plasma Protein Binding (PPB): | 95.65% |
| Volume Distribution (VD): | 1.196 | Fu: | 5.03% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.951 | CYP1A2-substrate: | 0.52 |
| CYP2C19-inhibitor: | 0.611 | CYP2C19-substrate: | 0.457 |
| CYP2C9-inhibitor: | 0.384 | CYP2C9-substrate: | 0.899 |
| CYP2D6-inhibitor: | 0.095 | CYP2D6-substrate: | 0.656 |
| CYP3A4-inhibitor: | 0.401 | CYP3A4-substrate: | 0.132 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.688 | Half-life (T1/2): | 0.279 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.036 | Human Hepatotoxicity (H-HT): | 0.014 |
| Drug-inuced Liver Injury (DILI): | 0.034 | AMES Toxicity: | 0.015 |
| Rat Oral Acute Toxicity: | 0.043 | Maximum Recommended Daily Dose: | 0.028 |
| Skin Sensitization: | 0.946 | Carcinogencity: | 0.13 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.988 |
| Respiratory Toxicity: | 0.355 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000455 |  |
0.903 | D0Z5BC |  |
0.561 | ||
| ENC000273 |  |
0.824 | D05ATI |  |
0.364 | ||
| ENC000510 |  |
0.757 | D0O1PH |  |
0.324 | ||
| ENC000475 |  |
0.700 | D0Z5SM |  |
0.323 | ||
| ENC000606 |  |
0.697 | D0O1TC |  |
0.319 | ||
| ENC000573 | 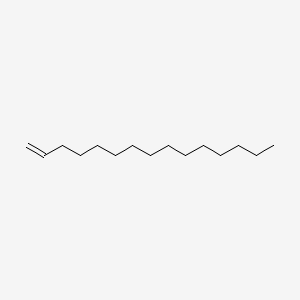 |
0.651 | D0E4WR | 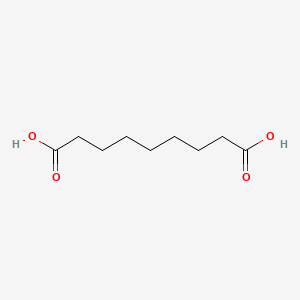 |
0.313 | ||
| ENC001139 | 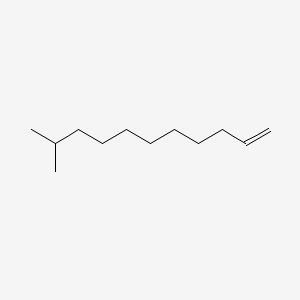 |
0.649 | D0Y8DP | 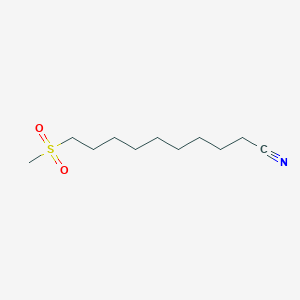 |
0.302 | ||
| ENC000267 |  |
0.639 | D0XN8C |  |
0.299 | ||
| ENC001684 |  |
0.639 | D03ZJE |  |
0.299 | ||
| ENC000425 |  |
0.609 | D0AY9Q |  |
0.296 | ||