NPs Basic Information
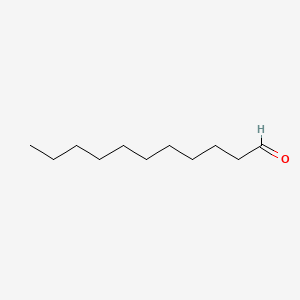
|
Name |
Undecanal
|
| Molecular Formula | C11H22O | |
| IUPAC Name* |
undecanal
|
|
| SMILES |
CCCCCCCCCCC=O
|
|
| InChI |
InChI=1S/C11H22O/c1-2-3-4-5-6-7-8-9-10-11-12/h11H,2-10H2,1H3
|
|
| InChIKey |
KMPQYAYAQWNLME-UHFFFAOYSA-N
|
|
| Synonyms |
UNDECANAL; 112-44-7; Undecanaldehyde; Undecylic aldehyde; n-Undecanal; Undecyl aldehyde; 1-Undecanal; Undecylaldehyde; Hendecanal; Hendecanaldehyde; n-Undecyl aldehyde; Aldehyde C-11; C11 aldehyde; C-11 aldehyde, undecylic; Aldehyde C-11, undecylic; FEMA No. 3092; NSC 22578; MFCD00007016; B6P0A9PSHN; CHEBI:46202; NSC-22578; Undecanone, alpha-; Undecanal (natural); EINECS 203-972-6; UNII-B6P0A9PSHN; BRN 1753213; AI3-05098; N-Indecyl aldehyde; Undecanal, 97%; 1gt4; aldehyde C11 undecylic; UNDECANAL [FCC]; UNDECANAL [FHFI]; DSSTox_CID_1688; EC 203-972-6; DSSTox_RID_76284; DSSTox_GSID_21688; SCHEMBL22333; Undecanal, FCC, >=96%; WLN: VH10; Undecanal, analytical standard; UNDECANONE, .ALPHA.-; CHEMBL1236576; DTXSID4021688; FEMA 3092; 1e02; NSC22578; ZINC1595727; Tox21_200538; LMFA06000064; AKOS009158017; CS-W004300; DB04093; NCGC00248685-01; NCGC00258092-01; CAS-112-44-7; SY048699; FT-0631645; U0009; UNDECANAL (ALDEHYDE C-11 UNDECYLIC); H10685; EN300-1721289; A894583; J-002779; Q7883008; Z993017862
|
|
| CAS | 112-44-7 | |
| PubChem CID | 8186 | |
| ChEMBL ID | CHEMBL1236576 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 170.29 | ALogp: | 4.3 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.37 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.604 | MDCK Permeability: | 0.00001790 |
| Pgp-inhibitor: | 0.059 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.994 |
| 30% Bioavailability (F30%): | 0.996 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.992 | Plasma Protein Binding (PPB): | 65.15% |
| Volume Distribution (VD): | 2.249 | Fu: | 16.83% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.801 | CYP1A2-substrate: | 0.395 |
| CYP2C19-inhibitor: | 0.401 | CYP2C19-substrate: | 0.171 |
| CYP2C9-inhibitor: | 0.26 | CYP2C9-substrate: | 0.885 |
| CYP2D6-inhibitor: | 0.081 | CYP2D6-substrate: | 0.203 |
| CYP3A4-inhibitor: | 0.118 | CYP3A4-substrate: | 0.083 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.573 | Half-life (T1/2): | 0.387 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.236 | Human Hepatotoxicity (H-HT): | 0.018 |
| Drug-inuced Liver Injury (DILI): | 0.057 | AMES Toxicity: | 0.069 |
| Rat Oral Acute Toxicity: | 0.03 | Maximum Recommended Daily Dose: | 0.026 |
| Skin Sensitization: | 0.968 | Carcinogencity: | 0.323 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.976 |
| Respiratory Toxicity: | 0.964 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000277 | 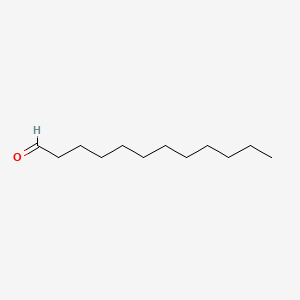 |
0.919 | D05ATI |  |
0.500 | ||
| ENC000267 | 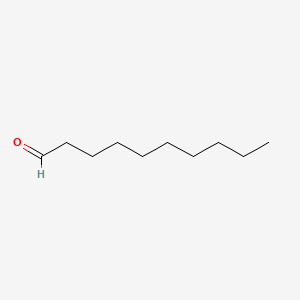 |
0.912 | D0Z5BC | 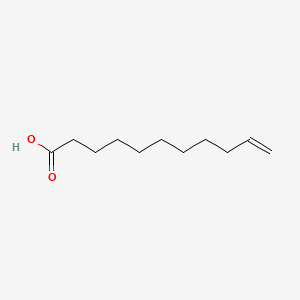 |
0.458 | ||
| ENC000606 | 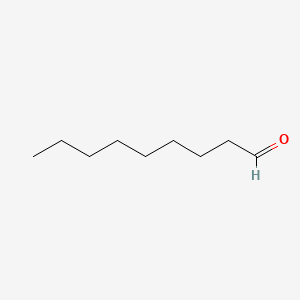 |
0.824 | D0Z5SM |  |
0.443 | ||
| ENC000607 | 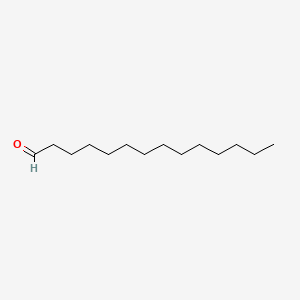 |
0.791 | D0Y8DP | 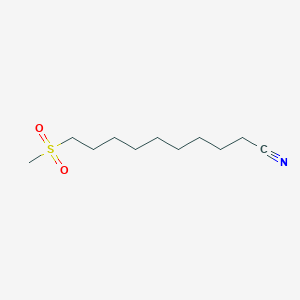 |
0.442 | ||
| ENC000273 | 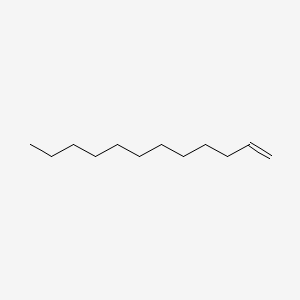 |
0.744 | D0O1PH |  |
0.429 | ||
| ENC000032 |  |
0.735 | D07ILQ |  |
0.403 | ||
| ENC000510 | 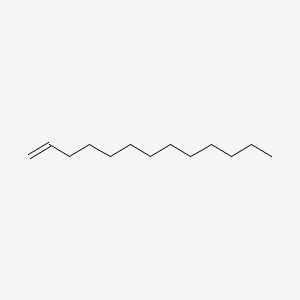 |
0.690 | D0XN8C | 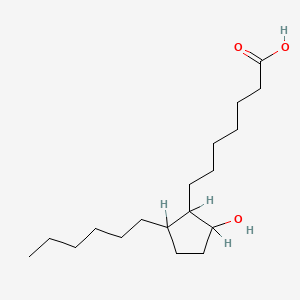 |
0.368 | ||
| ENC001656 | 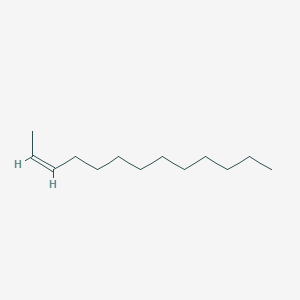 |
0.690 | D03ZJE | 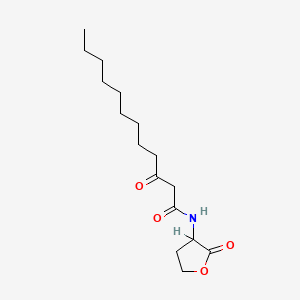 |
0.368 | ||
| ENC000473 |  |
0.667 | D05QNO | 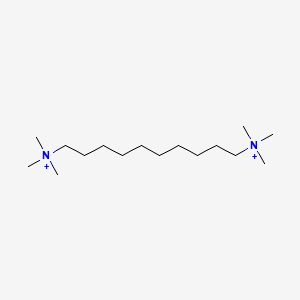 |
0.367 | ||
| ENC000502 | 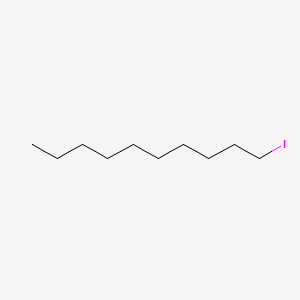 |
0.667 | D0O1TC | 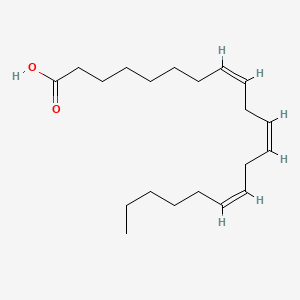 |
0.366 | ||