NPs Basic Information
|
Name |
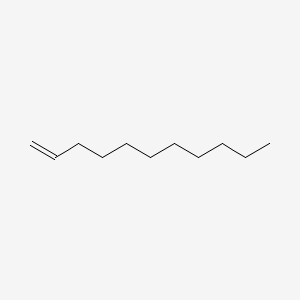
1-Undecene
|
| Molecular Formula | C11H22 | |
| IUPAC Name* |
undec-1-ene
|
|
| SMILES |
CCCCCCCCCC=C
|
|
| InChI |
InChI=1S/C11H22/c1-3-5-7-9-11-10-8-6-4-2/h3H,1,4-11H2,2H3
|
|
| InChIKey |
DCTOHCCUXLBQMS-UHFFFAOYSA-N
|
|
| Synonyms |
1-UNDECENE; 821-95-4; Undec-1-ene; n-1-Undecene; Undecene; 1-Hendecene; alpha-Undecene; alpha-Undecylene; alpha-Nonylethylene; Hendecene; Undecene-1; .alpha.-Undecene; A-Undecene; MFCD00008956; NSC-73983; 1446756A8F; CCRIS 5720; HSDB 1090; EINECS 212-483-7; NSC 73983; UNII-1446756A8F; 1-Undecene, 97%; .ALPHA.-UNDECYLENE; 1-UNDECENE [HSDB]; .ALPHA.-NONYLETHYLENE; DTXSID5061168; CHEBI:77444; NSC73983; ZINC1699445; EINECS 271-214-1; LMFA11000332; STL453737; AKOS009156849; LS-14020; DB-056580; FT-0608327; U0025; U0052; D92764; EC 271-214-1; Q14745306
|
|
| CAS | 821-95-4 | |
| PubChem CID | 13190 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.29 | ALogp: | 6.2 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 11 | QED Weighted: | 0.346 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.49 | MDCK Permeability: | 0.00001590 |
| Pgp-inhibitor: | 0.015 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.284 |
| 30% Bioavailability (F30%): | 0.613 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.98 | Plasma Protein Binding (PPB): | 96.88% |
| Volume Distribution (VD): | 1.367 | Fu: | 3.54% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.936 | CYP1A2-substrate: | 0.411 |
| CYP2C19-inhibitor: | 0.62 | CYP2C19-substrate: | 0.339 |
| CYP2C9-inhibitor: | 0.377 | CYP2C9-substrate: | 0.91 |
| CYP2D6-inhibitor: | 0.118 | CYP2D6-substrate: | 0.595 |
| CYP3A4-inhibitor: | 0.475 | CYP3A4-substrate: | 0.118 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.219 | Half-life (T1/2): | 0.224 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.045 | Human Hepatotoxicity (H-HT): | 0.013 |
| Drug-inuced Liver Injury (DILI): | 0.036 | AMES Toxicity: | 0.014 |
| Rat Oral Acute Toxicity: | 0.039 | Maximum Recommended Daily Dose: | 0.03 |
| Skin Sensitization: | 0.949 | Carcinogencity: | 0.107 |
| Eye Corrosion: | 0.993 | Eye Irritation: | 0.985 |
| Respiratory Toxicity: | 0.361 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000273 |  |
0.912 | D0Z5BC |  |
0.634 | ||
| ENC000460 |  |
0.903 | D05ATI |  |
0.418 | ||
| ENC000510 | 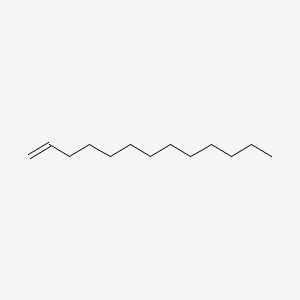 |
0.838 | D0Z5SM |  |
0.371 | ||
| ENC000475 | 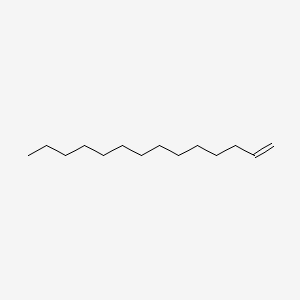 |
0.775 | D0O1PH |  |
0.366 | ||
| ENC000267 | 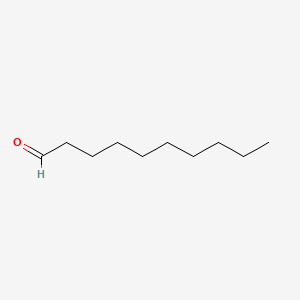 |
0.722 | D0Y8DP | 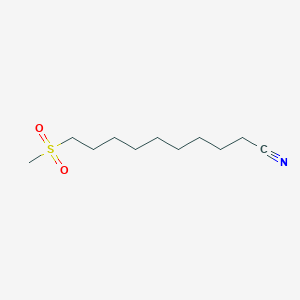 |
0.358 | ||
| ENC000573 |  |
0.721 | D03ZJE |  |
0.343 | ||
| ENC000425 |  |
0.674 | D0O1TC |  |
0.343 | ||
| ENC000275 |  |
0.667 | D07ILQ |  |
0.338 | ||
| ENC000493 |  |
0.639 | D0XN8C |  |
0.324 | ||
| ENC000542 |  |
0.639 | D05QNO |  |
0.317 | ||