NPs Basic Information
|
Name |
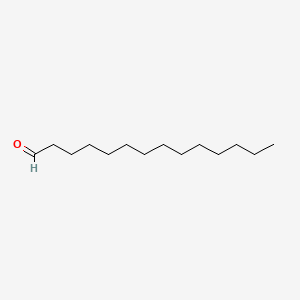
Tetradecanal
|
| Molecular Formula | C14H28O | |
| IUPAC Name* |
tetradecanal
|
|
| SMILES |
CCCCCCCCCCCCCC=O
|
|
| InChI |
InChI=1S/C14H28O/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15/h14H,2-13H2,1H3
|
|
| InChIKey |
UHUFTBALEZWWIH-UHFFFAOYSA-N
|
|
| Synonyms |
TETRADECANAL; Myristaldehyde; 124-25-4; Myristic aldehyde; n-Tetradecanal; Tetradecyl aldehyde; 1-Tetradecanal; Tetradecylaldehyde; Myristylaldehyde; Aldehyde C-14; C-14 aldehyde, myristic; Aldehyde C-14, myristic; Myristyl Aldehyde; n-Tetradecyl aldehyde; 1-Tetradecyl aldehyde; FEMA No. 2763; NSC 66435; 44AJ2LT15N; CHEBI:84067; FEMA 2763; NSC66435; NSC-66435; EINECS 204-692-7; BRN 1765987; UNII-44AJ2LT15N; AI3-36199; Tetradecanaldehyde; tetradecane aldehyde; 1la3; aldehyde C-14 myristic; DSSTox_CID_1665; TETRADECANAL, 98%; DSSTox_RID_76273; MYRISTALDEHYDE [FCC]; n-C13H27CHO; DSSTox_GSID_21665; SCHEMBL18604; MYRISTALDEHYDE [FHFI]; 4-01-00-03389 (Beilstein Handbook Reference); WLN: VH13; CH3(CH2)12CHO; PEACH ALDEHYDE (C14); CHEMBL2228569; DTXSID1021665; AMY6110; ZINC1693894; Tox21_202794; BBL102211; LMFA06000078; MFCD00007019; STL556010; AHHY-124-25-4; AKOS009158344; ZINC585138970; CS-W004303; GS-5775; NCGC00260340-01; 511542-15-7; CAS-124-25-4; FT-0674955; T2696; H10276; 124F254; A805213; Q6948297; W-108409
|
|
| CAS | 124-25-4 | |
| PubChem CID | 31291 | |
| ChEMBL ID | CHEMBL2228569 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 212.37 | ALogp: | 6.0 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 12 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 15 | QED Weighted: | 0.318 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.725 | MDCK Permeability: | 0.00001530 |
| Pgp-inhibitor: | 0.015 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.99 |
| 30% Bioavailability (F30%): | 0.999 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.971 | Plasma Protein Binding (PPB): | 87.04% |
| Volume Distribution (VD): | 2.906 | Fu: | 3.85% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.51 | CYP1A2-substrate: | 0.212 |
| CYP2C19-inhibitor: | 0.453 | CYP2C19-substrate: | 0.071 |
| CYP2C9-inhibitor: | 0.218 | CYP2C9-substrate: | 0.918 |
| CYP2D6-inhibitor: | 0.184 | CYP2D6-substrate: | 0.134 |
| CYP3A4-inhibitor: | 0.228 | CYP3A4-substrate: | 0.059 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.76 | Half-life (T1/2): | 0.211 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.311 | Human Hepatotoxicity (H-HT): | 0.015 |
| Drug-inuced Liver Injury (DILI): | 0.094 | AMES Toxicity: | 0.03 |
| Rat Oral Acute Toxicity: | 0.023 | Maximum Recommended Daily Dose: | 0.029 |
| Skin Sensitization: | 0.973 | Carcinogencity: | 0.173 |
| Eye Corrosion: | 0.994 | Eye Irritation: | 0.96 |
| Respiratory Toxicity: | 0.965 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000277 |  |
0.860 | D0Z5SM |  |
0.590 | ||
| ENC000573 |  |
0.792 | D05ATI |  |
0.579 | ||
| ENC000275 |  |
0.791 | D07ILQ |  |
0.537 | ||
| ENC000425 |  |
0.745 | D0O1PH |  |
0.514 | ||
| ENC001666 |  |
0.736 | D00AOJ |  |
0.467 | ||
| ENC001240 |  |
0.729 | D00FGR | 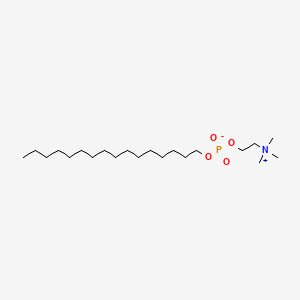 |
0.439 | ||
| ENC000475 | 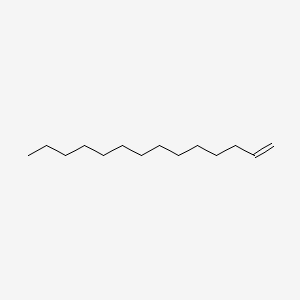 |
0.729 | D0P1RL | 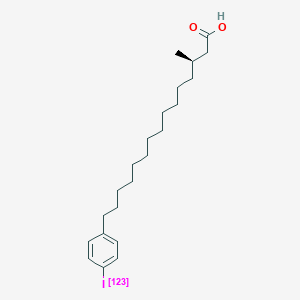 |
0.390 | ||
| ENC000422 | 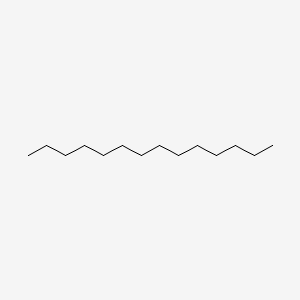 |
0.729 | D0Z5BC | 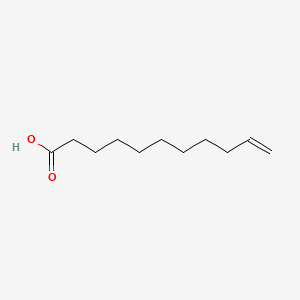 |
0.386 | ||
| ENC000267 | 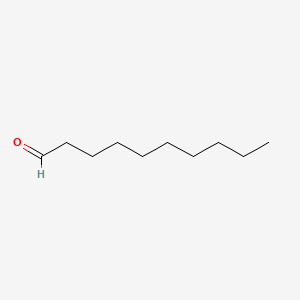 |
0.721 | D05QNO | 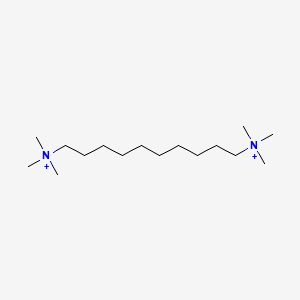 |
0.379 | ||
| ENC001686 | 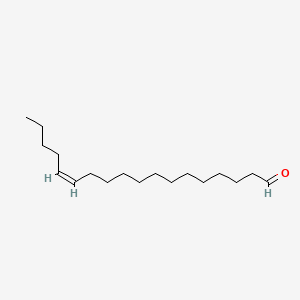 |
0.719 | D0Y8DP | 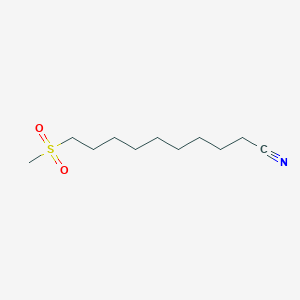 |
0.377 | ||