NPs Basic Information
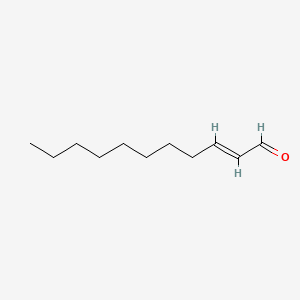
|
Name |
2-Undecenal
|
| Molecular Formula | C11H20O | |
| IUPAC Name* |
(E)-undec-2-enal
|
|
| SMILES |
CCCCCCCC/C=C/C=O
|
|
| InChI |
InChI=1S/C11H20O/c1-2-3-4-5-6-7-8-9-10-11-12/h9-11H,2-8H2,1H3/b10-9+
|
|
| InChIKey |
PANBRUWVURLWGY-MDZDMXLPSA-N
|
|
| Synonyms |
trans-2-Undecenal; 2-UNDECENAL; (E)-Undec-2-enal; 53448-07-0; 2463-77-6; (E)-2-undecenal; 2-Undecenal, (2E)-; (2E)-2-Undecenal; trans-2-Undecen-1-al; 2-Undecen-1-al; 2-Undecenal, (E)-; Undec-2-enal; 1337-83-3; trans-undec-2-enal; (2E)-undec-2-enal; FNP3S9MG30; 2-UNDECENAL (HIGH TRANS); Aldehyde iso C-11; UNII-F851M0LYFD; UNII-FNP3S9MG30; UNII-090E982ABR; FEMA No. 3423; (2e)-undecenal; undecenal (2e-); 2(E)-Undecenal; EINECS 215-656-5; EINECS 219-564-6; EINECS 258-559-3; 2-Undecenal, E-; 2-Undecenal, trans; Undec-2(E)-enal; AI3-36265; (e)-2-undecen-1-al; F851M0LYFD; CHEMBL451328; FEMA 3423; DTXSID20904470; CHEBI:132843; CHEBI:133173; 090E982ABR; trans-2-Undecenal, >=95%, FG; ZINC1849946; FEMA NO. 3423, E-; LMFA06000065; MFCD00014680; AKOS015901935; ZINC585138973; LS-14008; U0046; A817413; Q27278090; 2-Undecenal (High trans) stabilized with alpha-tocopherol
|
|
| CAS | 53448-07-0 | |
| PubChem CID | 5283356 | |
| ChEMBL ID | CHEMBL451328 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 168.28 | ALogp: | 4.2 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.301 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.436 | MDCK Permeability: | 0.00002880 |
| Pgp-inhibitor: | 0.003 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.802 |
| 30% Bioavailability (F30%): | 0.974 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.994 | Plasma Protein Binding (PPB): | 89.53% |
| Volume Distribution (VD): | 0.923 | Fu: | 9.81% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.96 | CYP1A2-substrate: | 0.578 |
| CYP2C19-inhibitor: | 0.746 | CYP2C19-substrate: | 0.715 |
| CYP2C9-inhibitor: | 0.433 | CYP2C9-substrate: | 0.967 |
| CYP2D6-inhibitor: | 0.112 | CYP2D6-substrate: | 0.828 |
| CYP3A4-inhibitor: | 0.116 | CYP3A4-substrate: | 0.128 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.111 | Half-life (T1/2): | 0.424 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.052 | Human Hepatotoxicity (H-HT): | 0.041 |
| Drug-inuced Liver Injury (DILI): | 0.087 | AMES Toxicity: | 0.498 |
| Rat Oral Acute Toxicity: | 0.037 | Maximum Recommended Daily Dose: | 0.054 |
| Skin Sensitization: | 0.958 | Carcinogencity: | 0.571 |
| Eye Corrosion: | 0.983 | Eye Irritation: | 0.983 |
| Respiratory Toxicity: | 0.951 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001599 |  |
0.912 | D0O1TC |  |
0.386 | ||
| ENC001598 |  |
0.824 | D0Z5BC | 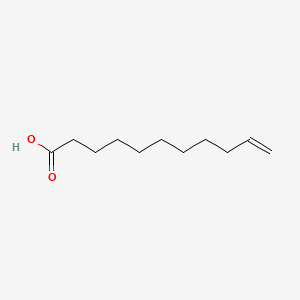 |
0.373 | ||
| ENC001597 | 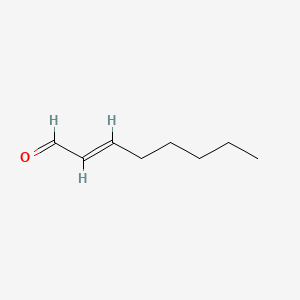 |
0.735 | D0O1PH |  |
0.370 | ||
| ENC001724 | 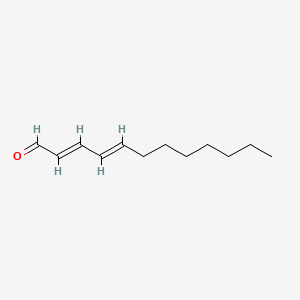 |
0.732 | D0UE9X | 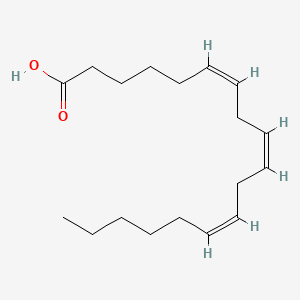 |
0.358 | ||
| ENC000606 | 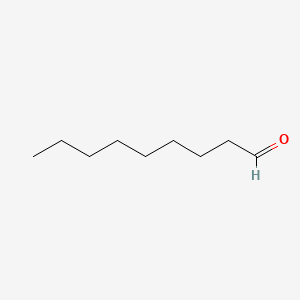 |
0.676 | D05ATI |  |
0.350 | ||
| ENC001684 | 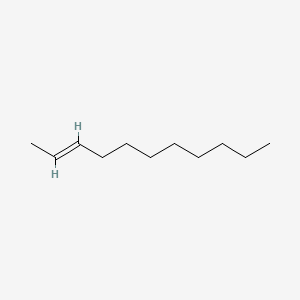 |
0.667 | D0OR6A | 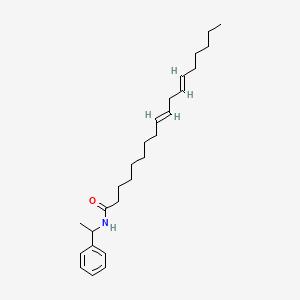 |
0.322 | ||
| ENC000267 | 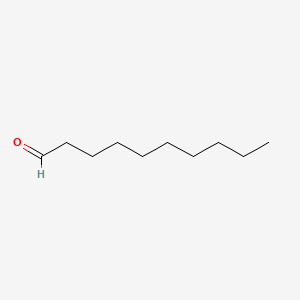 |
0.625 | D0Z5SM |  |
0.313 | ||
| ENC001654 | 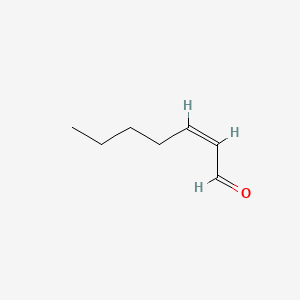 |
0.600 | D0E4WR | 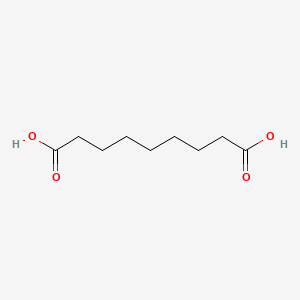 |
0.302 | ||
| ENC000032 |  |
0.595 | D09SRR | 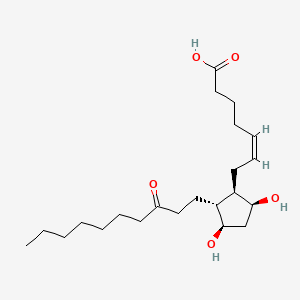 |
0.298 | ||
| ENC000460 | 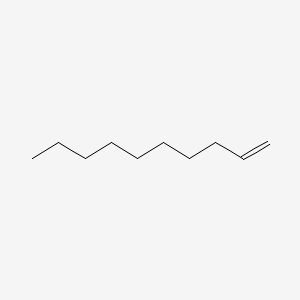 |
0.590 | D0Y8DP | 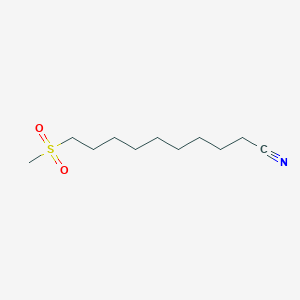 |
0.293 | ||