NPs Basic Information
|
Name |
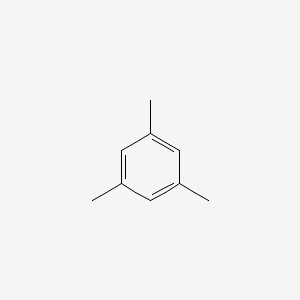
Mesitylene
|
| Molecular Formula | C9H12 | |
| IUPAC Name* |
1,3,5-trimethylbenzene
|
|
| SMILES |
CC1=CC(=CC(=C1)C)C
|
|
| InChI |
InChI=1S/C9H12/c1-7-4-8(2)6-9(3)5-7/h4-6H,1-3H3
|
|
| InChIKey |
AUHZEENZYGFFBQ-UHFFFAOYSA-N
|
|
| Synonyms |
MESITYLENE; 1,3,5-Trimethylbenzene; 108-67-8; sym-Trimethylbenzene; Benzene, 1,3,5-trimethyl-; 3,5-Dimethyltoluene; Fleet-X; Trimethylbenzol; s-Trimethylbenzene; 2,4,6-trimethylbenzene; 1,3,5-trimethyl-benzene; NSC 9273; Trimethylbenzene, 1,3,5-; CHEBI:34833; 887L18KQ6X; NSC-9273; DSSTox_CID_6797; DSSTox_RID_78217; DSSTox_GSID_26797; CAS-108-67-8; HSDB 92; EINECS 203-604-4; UN2325; UNII-887L18KQ6X; AI3-23973; CCRIS 8147; Mesitylene, 98%; MESITYLENE [MI]; MESITYLENE [HSDB]; MESITYLENE [INCI]; 1,3, 5-Trimethylbenzene; EC 203-604-4; BIDD:ER0286; Mesitylene (ACD/Name 4.0); Mesitylene, analytical standard; CHEMBL1797281; DTXSID6026797; WLN: 1R C1 E1; Mesitylene, reagent grade, 97%; NSC9273; BENZENE,1,3,5-TRIMETHYL; STR03436; ZINC1699890; Tox21_201452; Tox21_300341; MFCD00008538; STL268905; 1,3,5-Trimethylbenzene (Mesitylene); AKOS000120144; Mesitylene, purum, >=98.0% (GC); UN 2325; NCGC00247999-01; NCGC00247999-02; NCGC00254430-01; NCGC00259003-01; FT-0606520; S0658; T0470; EN300-19371; A801911; Q425161; 1,3,5-Trimethylbenzene 100 microg/mL in Methanol; J-002179; J-521685; 1,3,5-Trimethylbenzene [UN2325] [Flammable liquid]; F0001-0175; Mesitylene, certified reference material, TraceCERT(R)
|
|
| CAS | 108-67-8 | |
| PubChem CID | 7947 | |
| ChEMBL ID | CHEMBL1797281 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 120.19 | ALogp: | 3.4 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 9 | QED Weighted: | 0.492 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.332 | MDCK Permeability: | 0.00002340 |
| Pgp-inhibitor: | 0.003 | Pgp-substrate: | 0.008 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.276 |
| 30% Bioavailability (F30%): | 0.851 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.984 | Plasma Protein Binding (PPB): | 89.20% |
| Volume Distribution (VD): | 0.766 | Fu: | 6.59% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.763 | CYP1A2-substrate: | 0.922 |
| CYP2C19-inhibitor: | 0.758 | CYP2C19-substrate: | 0.896 |
| CYP2C9-inhibitor: | 0.143 | CYP2C9-substrate: | 0.485 |
| CYP2D6-inhibitor: | 0.387 | CYP2D6-substrate: | 0.881 |
| CYP3A4-inhibitor: | 0.145 | CYP3A4-substrate: | 0.534 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.798 | Half-life (T1/2): | 0.758 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.035 | Human Hepatotoxicity (H-HT): | 0.172 |
| Drug-inuced Liver Injury (DILI): | 0.055 | AMES Toxicity: | 0.036 |
| Rat Oral Acute Toxicity: | 0.035 | Maximum Recommended Daily Dose: | 0.098 |
| Skin Sensitization: | 0.344 | Carcinogencity: | 0.247 |
| Eye Corrosion: | 0.973 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.091 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000692 | 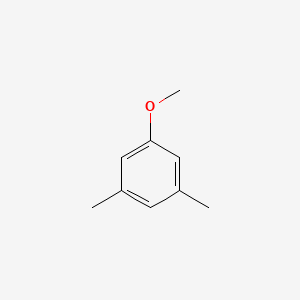 |
0.594 | D0S5CH | 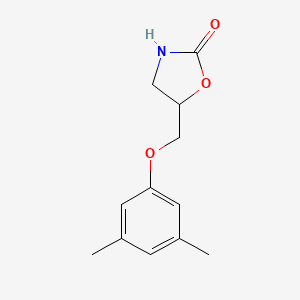 |
0.353 | ||
| ENC000180 | 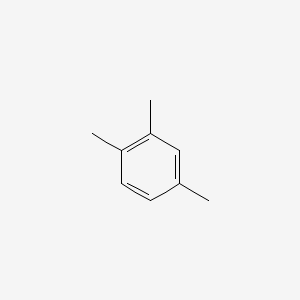 |
0.412 | D06GIP | 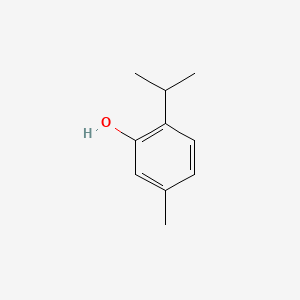 |
0.293 | ||
| ENC000239 | 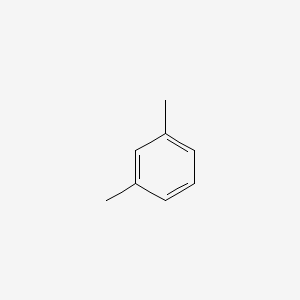 |
0.394 | D0FA2O | 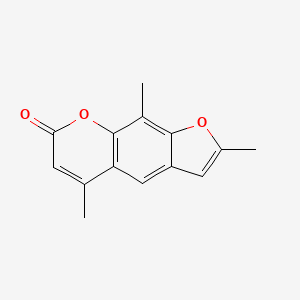 |
0.268 | ||
| ENC000736 | 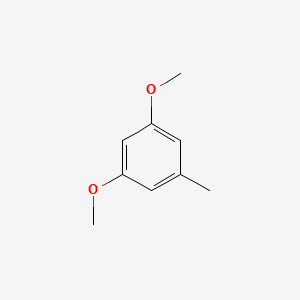 |
0.385 | D07EXH | 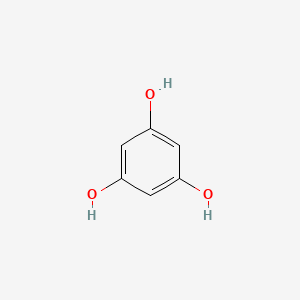 |
0.231 | ||
| ENC000353 | 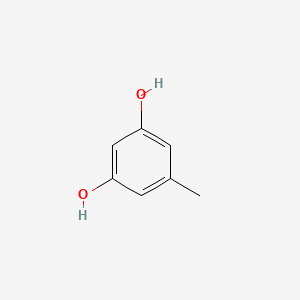 |
0.371 | D0X0RI | 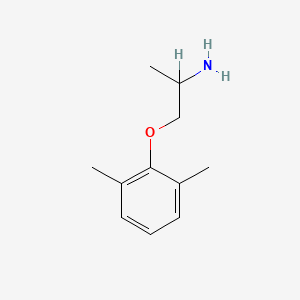 |
0.229 | ||
| ENC000491 | 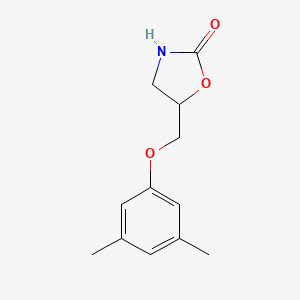 |
0.353 | D0M8RC | 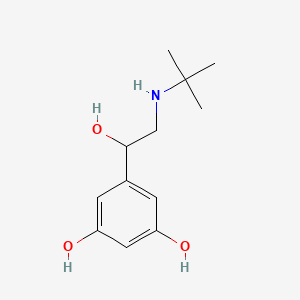 |
0.222 | ||
| ENC000181 | 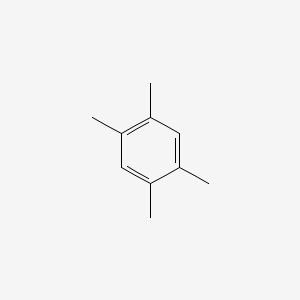 |
0.351 | D05VIX | 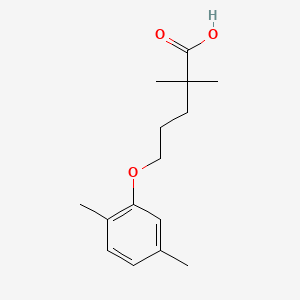 |
0.220 | ||
| ENC001186 | 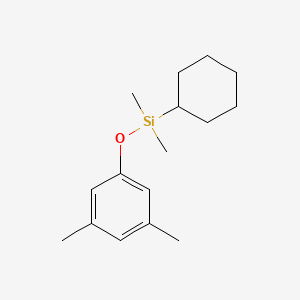 |
0.345 | D01PJR | 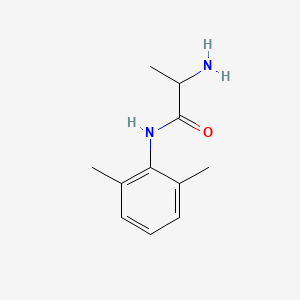 |
0.220 | ||
| ENC000498 | 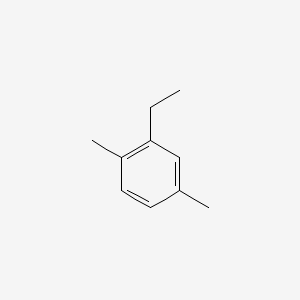 |
0.342 | D02UFG | 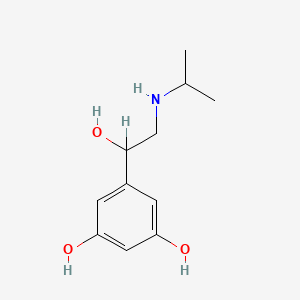 |
0.208 | ||
| ENC000364 |  |
0.333 | D05YBZ | 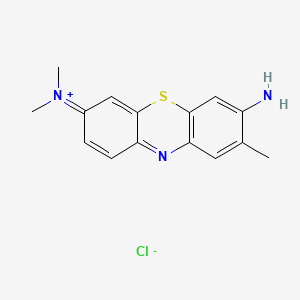 |
0.200 | ||