NPs Basic Information
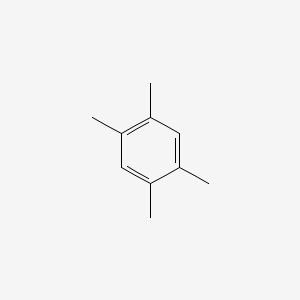
|
Name |
1,2,4,5-Tetramethylbenzene
|
| Molecular Formula | C10H14 | |
| IUPAC Name* |
1,2,4,5-tetramethylbenzene
|
|
| SMILES |
CC1=CC(=C(C=C1C)C)C
|
|
| InChI |
InChI=1S/C10H14/c1-7-5-9(3)10(4)6-8(7)2/h5-6H,1-4H3
|
|
| InChIKey |
SQNZJJAZBFDUTD-UHFFFAOYSA-N
|
|
| Synonyms |
1,2,4,5-TETRAMETHYLBENZENE; 95-93-2; Durene; Durol; Benzene, 1,2,4,5-tetramethyl-; p-Xylene, 2,5-dimethyl-; 1,2,4,5-tetramethyl-benzene; CHEBI:38978; 181426CFYB; NSC-6770; DSSTox_CID_9124; DSSTox_RID_78675; DSSTox_GSID_29124; CAS-95-93-2; NSC 6770; EINECS 202-465-7; Duren; UNII-181426CFYB; AI3-25182; CCRIS 8660; 2,5-dimethyl-p-xylene; DURENE [MI]; NCIMech_000514; BIDD:ER0685; 1,2,4,5-tetramethyl benzene; CHEMBL1797134; DTXSID1029124; NSC6770; WLN: 1R B1 D1 E1; ZINC1295393; Tox21_201920; Tox21_303511; 1,2,4,5-TETRAMETHLYBENZENE; MFCD00008528; STL268879; 1,2,4,5-Tetramethylbenzene, 98%; AKOS000119934; NCGC00249135-01; NCGC00257264-01; NCGC00259469-01; LS-13806; DB-038211; FT-0625629; T0140; T0714; 1,2,4,5-TETRAMETHYLBENZENE (DURENE); EN300-19396; Q907919; W-100151; F0001-2285; Z104473710; 1,2,4,5-Tetramethylbenzene, Standard for quantitative NMR, TraceCERT(R)
|
|
| CAS | 95-93-2 | |
| PubChem CID | 7269 | |
| ChEMBL ID | CHEMBL1797134 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 134.22 | ALogp: | 4.0 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.507 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.631 | MDCK Permeability: | 0.00001950 |
| Pgp-inhibitor: | 0.079 | Pgp-substrate: | 0.02 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.962 |
| 30% Bioavailability (F30%): | 0.968 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.899 | Plasma Protein Binding (PPB): | 92.28% |
| Volume Distribution (VD): | 0.914 | Fu: | 6.03% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.762 | CYP1A2-substrate: | 0.948 |
| CYP2C19-inhibitor: | 0.603 | CYP2C19-substrate: | 0.898 |
| CYP2C9-inhibitor: | 0.13 | CYP2C9-substrate: | 0.717 |
| CYP2D6-inhibitor: | 0.302 | CYP2D6-substrate: | 0.933 |
| CYP3A4-inhibitor: | 0.11 | CYP3A4-substrate: | 0.523 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.843 | Half-life (T1/2): | 0.429 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.076 |
| Drug-inuced Liver Injury (DILI): | 0.068 | AMES Toxicity: | 0.221 |
| Rat Oral Acute Toxicity: | 0.023 | Maximum Recommended Daily Dose: | 0.619 |
| Skin Sensitization: | 0.249 | Carcinogencity: | 0.656 |
| Eye Corrosion: | 0.961 | Eye Irritation: | 0.996 |
| Respiratory Toxicity: | 0.036 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001026 | 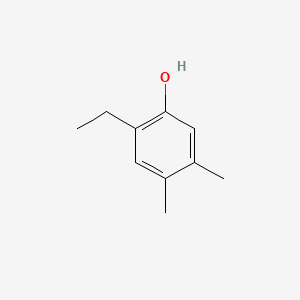 |
0.447 | D0Y4DY | 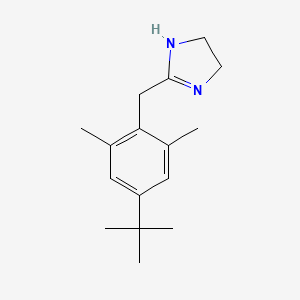 |
0.271 | ||
| ENC000342 | 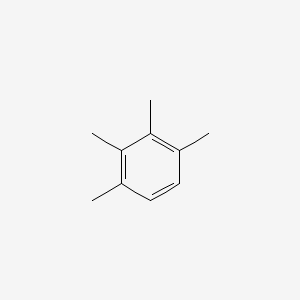 |
0.444 | D0FA2O | 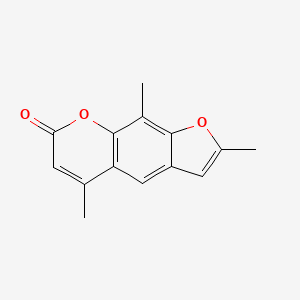 |
0.259 | ||
| ENC000180 | 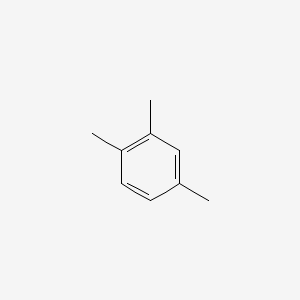 |
0.429 | D0X0RI | 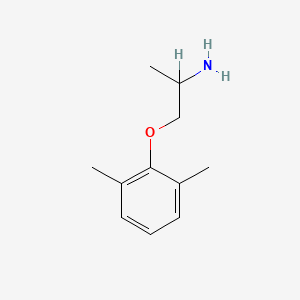 |
0.245 | ||
| ENC000728 | 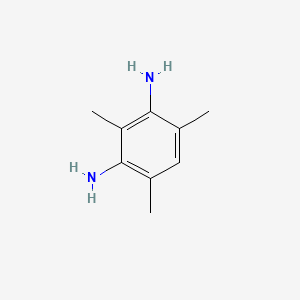 |
0.385 | D01PJR | 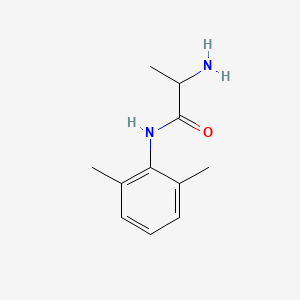 |
0.235 | ||
| ENC000084 | 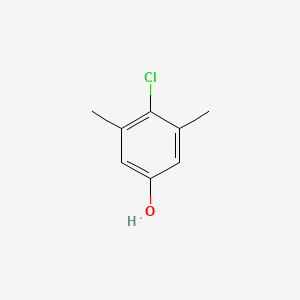 |
0.368 | D06GIP | 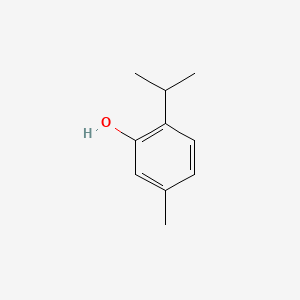 |
0.222 | ||
| ENC000614 |  |
0.351 | D0X4RN | 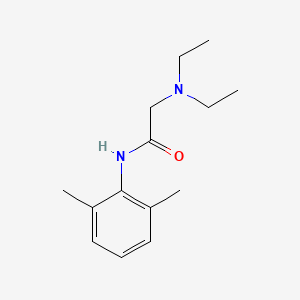 |
0.220 | ||
| ENC000364 |  |
0.351 | D0U3DU | 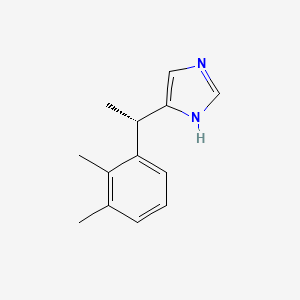 |
0.214 | ||
| ENC000242 | 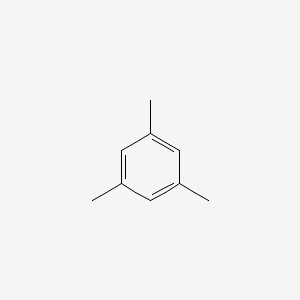 |
0.351 | D05VIX | 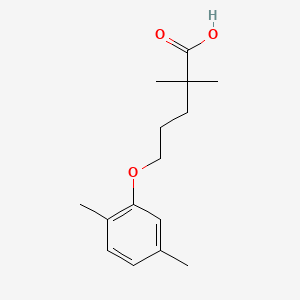 |
0.213 | ||
| ENC002336 | 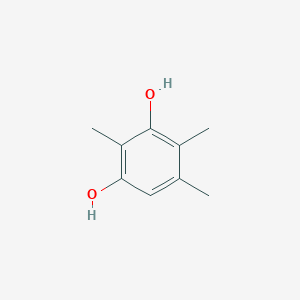 |
0.317 | D0S5CH | 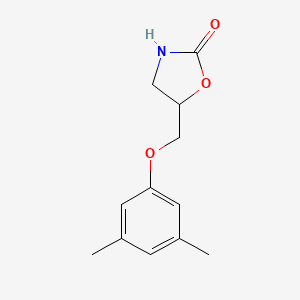 |
0.203 | ||
| ENC005230 | 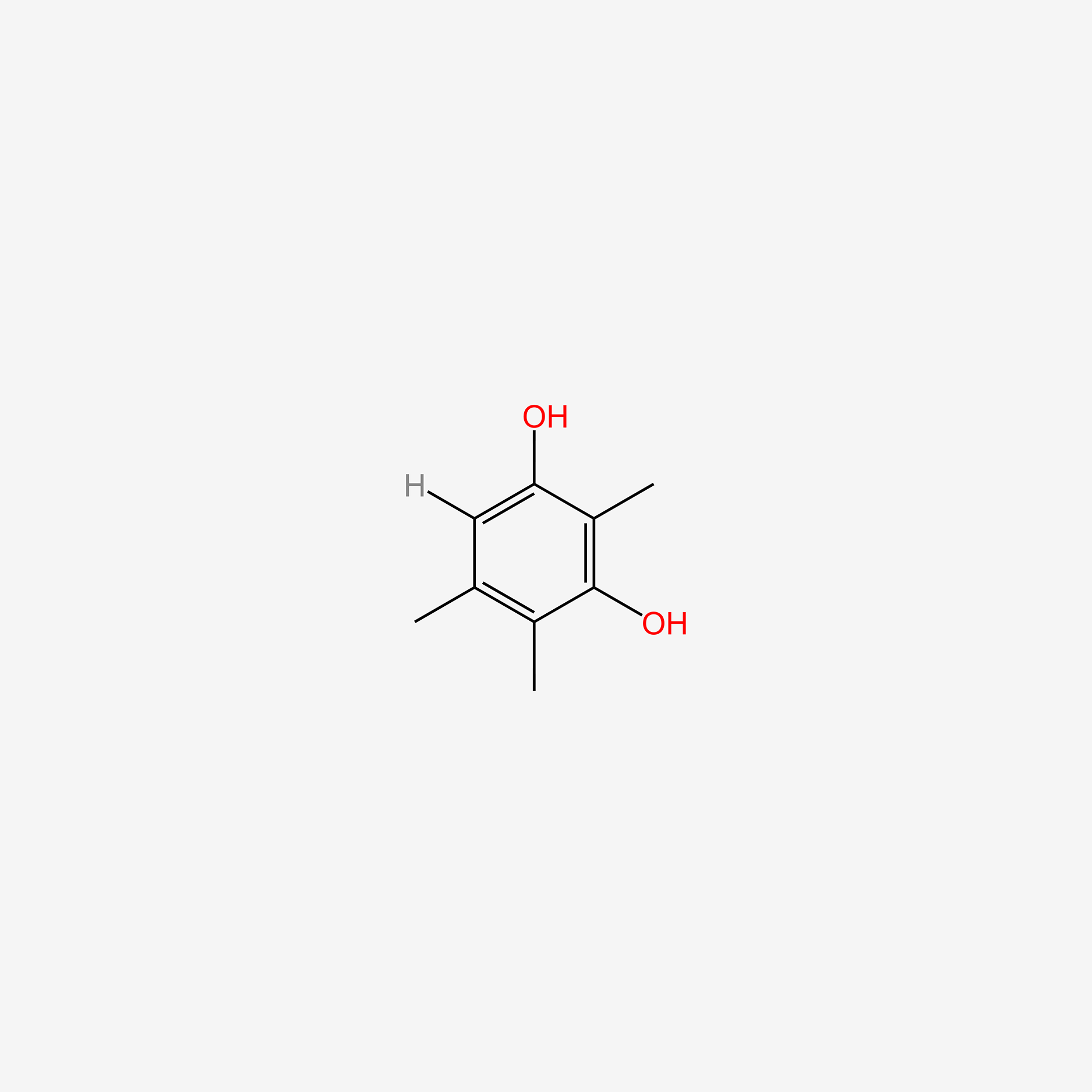 |
0.317 | D09EBS | 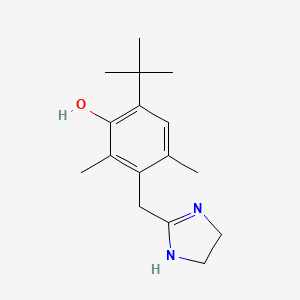 |
0.203 | ||