NPs Basic Information
|
Name |
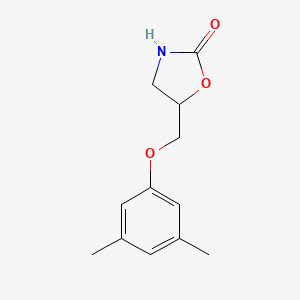
Metaxalone
|
| Molecular Formula | C12H15NO3 | |
| IUPAC Name* |
5-[(3,5-dimethylphenoxy)methyl]-1,3-oxazolidin-2-one
|
|
| SMILES |
CC1=CC(=CC(=C1)OCC2CNC(=O)O2)C
|
|
| InChI |
InChI=1S/C12H15NO3/c1-8-3-9(2)5-10(4-8)15-7-11-6-13-12(14)16-11/h3-5,11H,6-7H2,1-2H3,(H,13,14)
|
|
| InChIKey |
IMWZZHHPURKASS-UHFFFAOYSA-N
|
|
| Synonyms |
METAXALONE; 1665-48-1; Skelaxin; Zorane; Methaxalonum; Metaxalon; Methoxolone; 5-((3,5-Dimethylphenoxy)methyl)oxazolidin-2-one; Metazalone; Metazolone; Metaxolone; AHR-438; 5-[(3,5-Dimethylphenoxy)methyl]-1,3-oxazolidin-2-one; Metaxalonum [Latin]; Metassalone [DCIT]; 2-Oxazolidinone, 5-[(3,5-dimethylphenoxy)methyl]-; AHR 438; Metaxalonum [INN-Latin]; Metaxalona [INN-Spanish]; NSC 170959; 5-(3,5-Xyloloxymethyl)oxazolidin-2-one; CL 39,148; 5-[(3,5-Xylyloxy)methyl]-2-oxazolidinone; Flexura; 2-Oxazolidinone, 5-((3,5-xylyloxy)methyl)-; 5-((3,5-Dimethylphenoxy)methyl)-2-oxazolidinone; 5-((3,5-Xylyloxy)methyl)-2-oxazolidinone; MFCD00867700; NSC170959; 5-[(3,5-dimethylphenoxy)methyl]oxazolidin-2-one; NSC-170959; 2-Oxazolidinone, 5-((3,5-dimethylphenoxy)methyl)-; 5-(3,5-dimethylphenoxymethyl)-1,3-oxazolidin-2-one; MLS003106749; 1NMA9J598Y; CHEBI:6797; 5-[(3,5-Dimethylphenoxy)methyl]-2-oxazolidinone; NCGC00095116-01; DSSTox_CID_3269; 2-Oxazolidinone, 5-[(3,5-xylyloxy)methyl]-; DSSTox_RID_76949; DSSTox_GSID_23269; Metaxalona [Spanish]; Metassalone; Metaxalona; Metaxalonum; CAS-1665-48-1; Metaxalone [USAN:INN:BAN]; HSDB 3236; SR-05000001978; EINECS 216-777-6; 5-((3,5-Dimethylphenoxy)methyl)-1,3-oxazolidin-2-one; BRN 0884592; UNII-1NMA9J598Y; .meta.Xalone; .meta.Zalone; .meta.Zolone; .meta.Xalon; Skelaxin (TN); Spectrum_001741; METAXALONE [MI]; SpecPlus_000656; Metaxalone (USP/INN); METAXALONE [INN]; Spectrum2_000548; Spectrum3_001666; Spectrum4_000612; Spectrum5_001685; METAXALONE [HSDB]; METAXALONE [USAN]; METAXALONE [VANDF]; 5-(3,5-dimethylphenoxymethyl)-2-oxazolidinone; METAXALONE [MART.]; METAXALONE [USP-RS]; METAXALONE [WHO-DD]; Oprea1_438855; SCHEMBL34908; BSPBio_003451; KBioGR_001164; KBioSS_002221; DivK1c_006752; SPECTRUM1504229; SPBio_000595; AHR438; GTPL7609; CHEMBL1079604; DTXSID3023269; METAXALONE [ORANGE BOOK]; Metaxalone, >=98% (HPLC); KBio1_001696; KBio2_002221; KBio2_004789; KBio2_007357; KBio3_002671; HMS1922H07; HMS2093C22; METAXALONE [USP MONOGRAPH]; Pharmakon1600-01504229; BCP28377; HY-B0678; Tox21_111428; WLN: T5MVOTJ D1OR C1 E1; 2-Oxazolidinone,5-xylyloxy)methyl]-; CCG-39592; NSC758703; s3730; STL450994; STL451511; AKOS009035315; Tox21_111428_1; DB00660; FS-3218; NSC-758703; NCGC00095116-02; NCGC00095116-03; NCGC00095116-05; SMR001821638; SY052772; SBI-0052859.P002; 2-Oxazolidinone,5-dimethylphenoxy)methyl]-; DB-043656; AM20060525; FT-0603568; M2578; 5-(3,5-dimethylphenoxy)methyl-2-oxazolidinone; C07934; D00773; EN300-119839; AB00053284_04; 665M481; A810747; J-010295; Q6823309; SR-05000001978-1; SR-05000001978-3; BRD-A94709349-001-02-6; BRD-A94709349-001-03-4; Z106952910; 5-[(3,5-Dimethylphenoxy)methyl]-1,3-oxazolidin-2-one #; 5-{[(3,5-dimethylphenyl)oxy]methyl}-1,3-oxazolidin-2-one; 5-[(3,5-dimethylphenoxy)methyl]-4,5-dihydro-1,3-oxazol-2-ol; Metaxalone, United States Pharmacopeia (USP) Reference Standard; 105801-80-7
|
|
| CAS | 1665-48-1 | |
| PubChem CID | 15459 | |
| ChEMBL ID | CHEMBL1079604 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 221.25 | ALogp: | 2.2 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 47.6 | Aromatic Rings: | 2 |
| Heavy Atoms: | 16 | QED Weighted: | 0.852 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.391 | MDCK Permeability: | 0.00007690 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.044 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.975 | Plasma Protein Binding (PPB): | 86.32% |
| Volume Distribution (VD): | 0.604 | Fu: | 9.41% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.636 | CYP1A2-substrate: | 0.177 |
| CYP2C19-inhibitor: | 0.741 | CYP2C19-substrate: | 0.789 |
| CYP2C9-inhibitor: | 0.123 | CYP2C9-substrate: | 0.32 |
| CYP2D6-inhibitor: | 0.095 | CYP2D6-substrate: | 0.877 |
| CYP3A4-inhibitor: | 0.368 | CYP3A4-substrate: | 0.327 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.174 | Half-life (T1/2): | 0.789 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.047 | Human Hepatotoxicity (H-HT): | 0.696 |
| Drug-inuced Liver Injury (DILI): | 0.153 | AMES Toxicity: | 0.622 |
| Rat Oral Acute Toxicity: | 0.031 | Maximum Recommended Daily Dose: | 0.186 |
| Skin Sensitization: | 0.62 | Carcinogencity: | 0.829 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.06 |
| Respiratory Toxicity: | 0.039 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000692 | 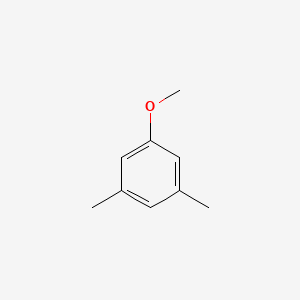 |
0.440 | D0S5CH | 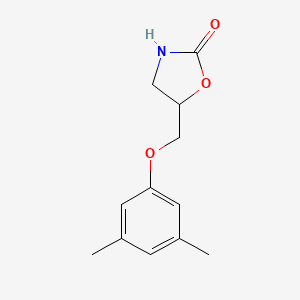 |
1.000 | ||
| ENC000242 |  |
0.353 | D0FA2O | 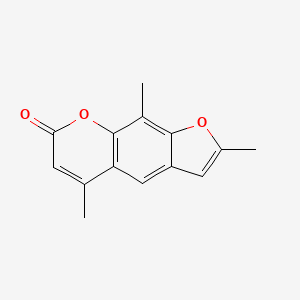 |
0.227 | ||
| ENC001186 | 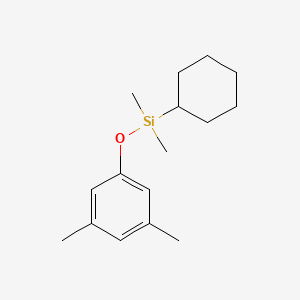 |
0.338 | D0V9JR | 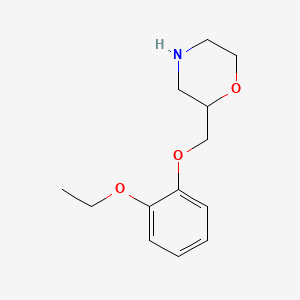 |
0.218 | ||
| ENC000736 | 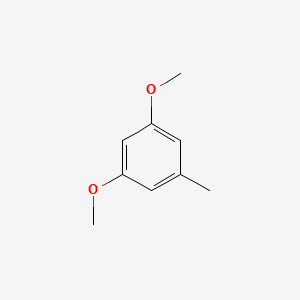 |
0.316 | D07TGY | 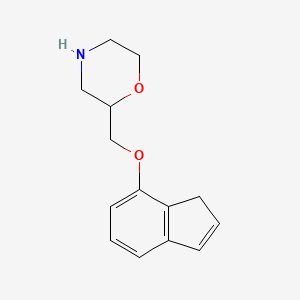 |
0.215 | ||
| ENC002445 | 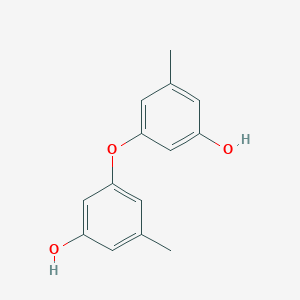 |
0.314 | D07GRH | 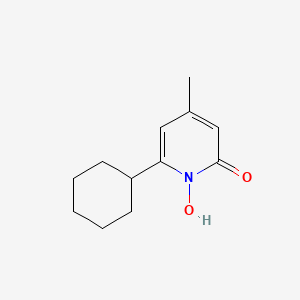 |
0.208 | ||
| ENC004151 | 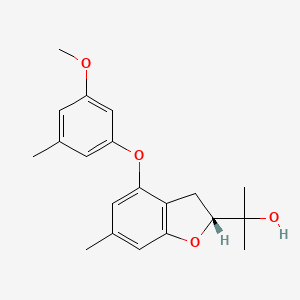 |
0.306 | D05VIX | 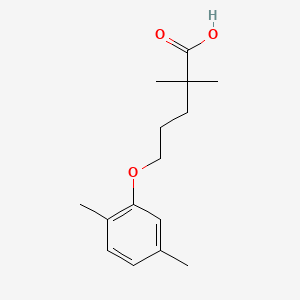 |
0.208 | ||
| ENC005186 | 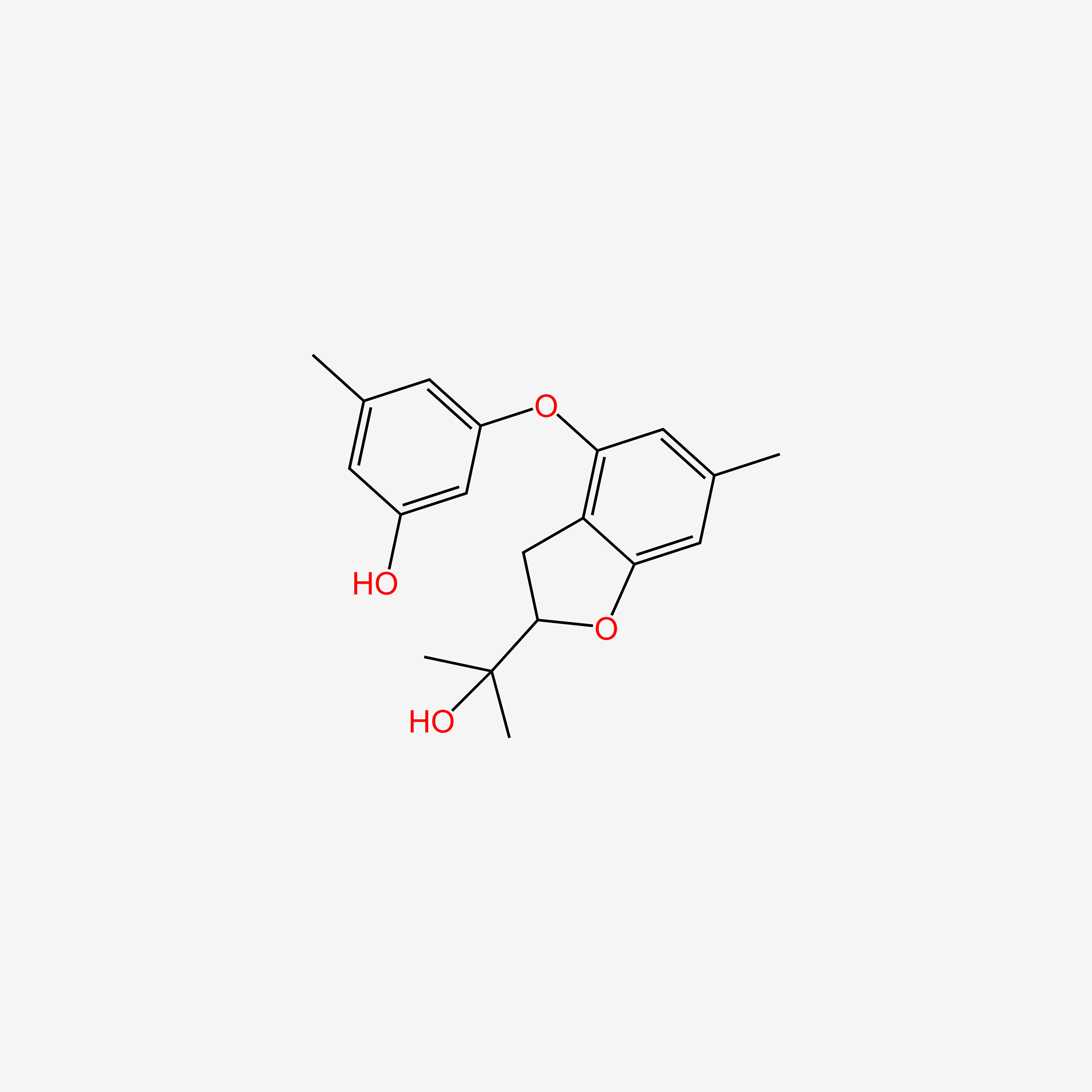 |
0.301 | D0NG7O | 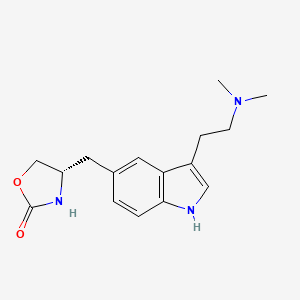 |
0.207 | ||
| ENC006021 | 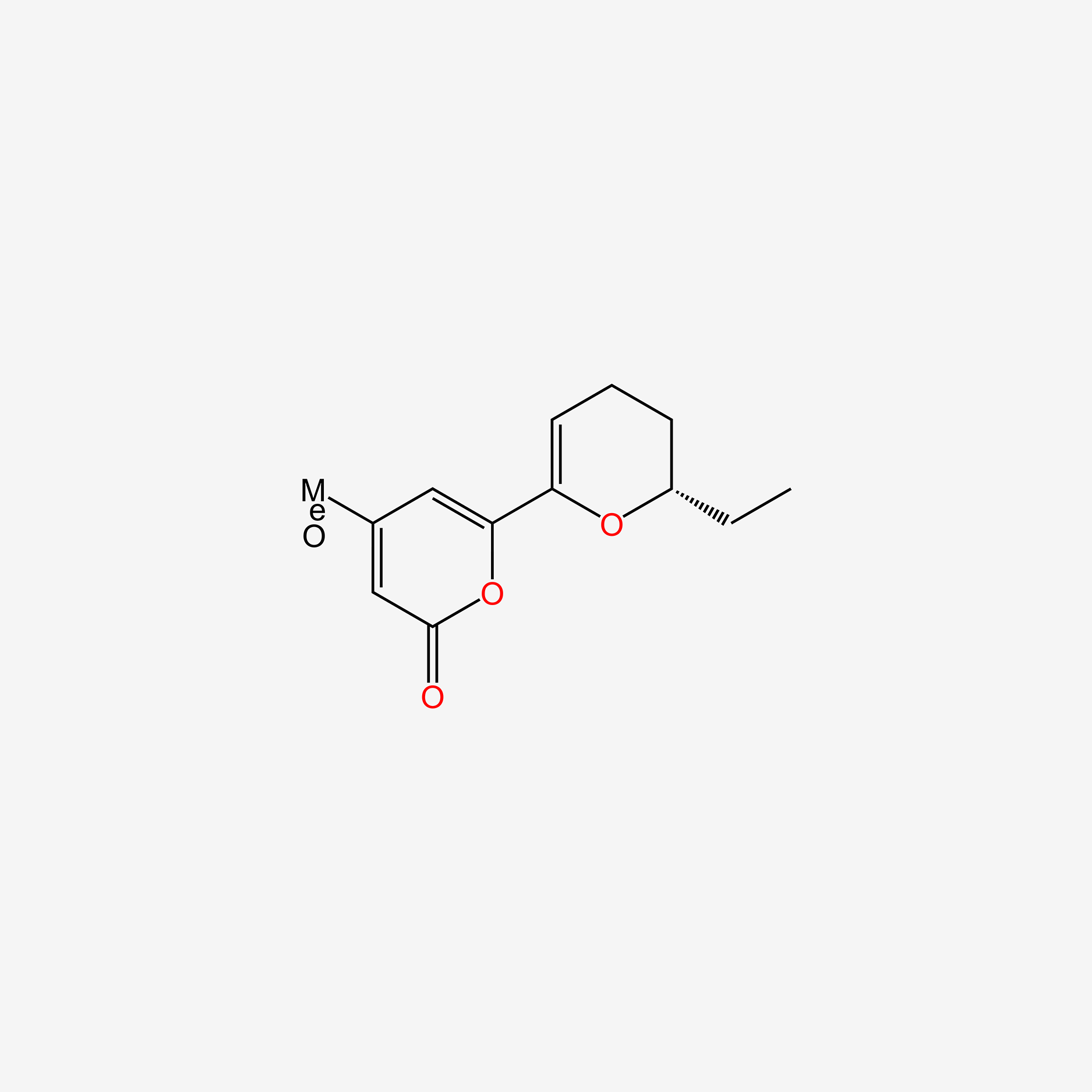 |
0.292 | D0Y4DY | 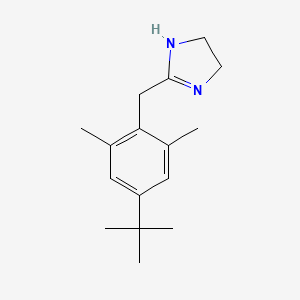 |
0.205 | ||
| ENC001355 | 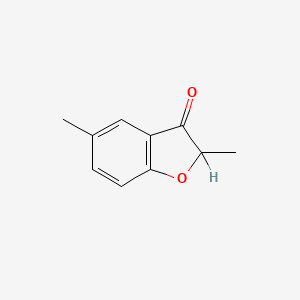 |
0.279 | D07MGA |  |
0.205 | ||
| ENC003683 | 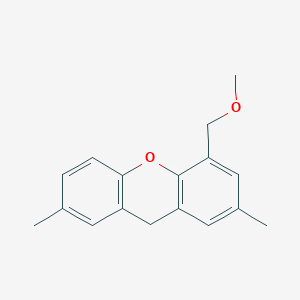 |
0.269 | D06GDY | 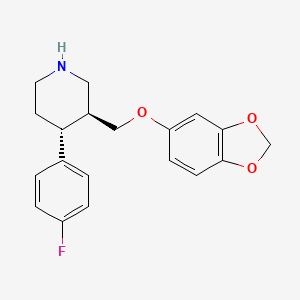 |
0.196 | ||