NPs Basic Information
|
Name |
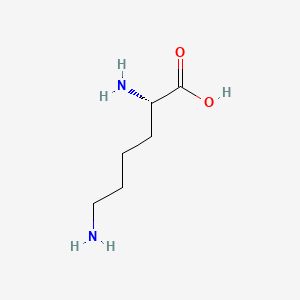
Lysine
|
| Molecular Formula | C6H14N2O2 | |
| IUPAC Name* |
(2S)-2,6-diaminohexanoic acid
|
|
| SMILES |
C(CCN)C[C@@H](C(=O)O)N
|
|
| InChI |
InChI=1S/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h5H,1-4,7-8H2,(H,9,10)/t5-/m0/s1
|
|
| InChIKey |
KDXKERNSBIXSRK-YFKPBYRVSA-N
|
|
| Synonyms |
L-lysine; lysine; 56-87-1; lysine acid; h-Lys-oh; (2S)-2,6-diaminohexanoic acid; (S)-Lysine; Aminutrin; L-(+)-Lysine; (S)-2,6-Diaminohexanoic acid; alpha-Lysine; Hydrolysin; Lysinum [Latin]; L-lys; Lisina [Spanish]; L-Norleucine, 6-amino-; Lysine, L-; Lysinum; (S)-alpha,epsilon-Diaminocaproic acid; Lysine [USAN:INN]; (S)-2,6-Diaminocaproic acid; LYS (IUPAC abbreviation); L-2,6-Diaminocaproic acid; 25104-18-1; Hexanoic acid, 2,6-diamino-, (S)-; lys; CHEBI:18019; a-Lysine; 2,6-Diaminohexanoic acid, (S)-; L-Lysin; BRN 1722531; AI3-26523; (+)-S-Lysine; 6-ammonio-L-norleucine; 12798-06-0; lysin; L-Lysine base; K3Z4F929H6; HSDB 2108; L-2,6-Diaminocaproate; Lisina; L-LYSINE, MONOACETATE; MFCD00064433; 3H-Lysine; 2,6-diaminohexanoate; EINECS 200-294-2; lysina; UNII-K3Z4F929H6; L-Lysine, labeled with tritium; Ketporofen lysine; .alpha.-Lysine; 1ozv; 1yxd; 3h-l-lysine; 6-amino-Aminutrin; NCGC00164527-01; H-Lys; (-)-lysine; 6-amino-L-Norleucine; a,e-Diaminocaproic acid; Lysine (USAN/INN); L-2,6-Diainohexanoate; LYSINE [VANDF]; LYSINE [HSDB]; LYSINE [INCI]; LYSINE [USAN]; LYSINE [INN]; L-LYSINE [FHFI]; LYSINE [WHO-DD]; (S)-a,e-Diaminocaproate; LYSINE [II]; LYSINE [MI]; LYSINE [MART.]; DSSTox_CID_3232; L-Lysine, >=97%; bmse000043; bmse000914; Epitope ID:136017; (S)-2,6-Diaminohexanoate; L-2,6-Diainohexanoic acid; CHEMBL8085; DSSTox_RID_76935; DSSTox_GSID_23232; GTPL724; (S)-2,6-diamino-Hexanoate; (S)-a,e-Diaminocaproic acid; 4-04-00-02717 (Beilstein Handbook Reference); L-Lysine, analytical standard; L-Lysine, >=98%, FG; DTXSID6023232; (S)-2,6-diamino-Hexanoic acid; L-Lysine, >=98% (TLC); BDBM217367; (2S)-2,6-Diamino-hexanoic acid; ACT02654; HY-N0469; L-H2N(CH2)4CH(NH2)COOH; ZINC1532522; Tox21_112158; Ethyl3,5-dichloro-4-propoxybenzoate; s5630; .alpha.,.epsilon.-Diaminocaproic acid; AKOS006239081; AKOS015855172; CCG-266180; CS-W019758; DB00123; CAS-56-87-1; NCGC00166296-02; 20166-34-1; AC-14492; AS-11733; TYROSINE IMPURITY B [EP IMPURITY]; (S)-.alpha.,.epsilon.-Diaminocaproic acid; L-Lysine, crystallized, >=98.0% (NT); AM20100376; L0129; L-Lysine, Vetec(TM) reagent grade, >=98%; A20652; C00047; D02304; 064L433; A904498; A919375; J-521651; (S)-2,6-Diaminocaproic acid;(S)-(+)-Lysine;Lysine; Q20816880; F0001-1472; 0013CD6B-1671-4369-B1BE-F531611E50C7
|
|
| CAS | 56-87-1 | |
| PubChem CID | 5962 | |
| ChEMBL ID | CHEMBL8085 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 146.19 | ALogp: | -3.0 |
| HBD: | 3 | HBA: | 4 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 89.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.475 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -6.156 | MDCK Permeability: | 0.00568378 |
| Pgp-inhibitor: | 0.013 | Pgp-substrate: | 0.013 |
| Human Intestinal Absorption (HIA): | 0.232 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.294 | Plasma Protein Binding (PPB): | 5.99% |
| Volume Distribution (VD): | 0.703 | Fu: | 93.95% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.011 | CYP1A2-substrate: | 0.045 |
| CYP2C19-inhibitor: | 0.03 | CYP2C19-substrate: | 0.058 |
| CYP2C9-inhibitor: | 0.008 | CYP2C9-substrate: | 0.118 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.288 |
| CYP3A4-inhibitor: | 0.009 | CYP3A4-substrate: | 0.036 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.617 | Half-life (T1/2): | 0.499 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.039 | Human Hepatotoxicity (H-HT): | 0.073 |
| Drug-inuced Liver Injury (DILI): | 0.008 | AMES Toxicity: | 0.262 |
| Rat Oral Acute Toxicity: | 0.185 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.527 | Carcinogencity: | 0.09 |
| Eye Corrosion: | 0.019 | Eye Irritation: | 0.088 |
| Respiratory Toxicity: | 0.377 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000137 | 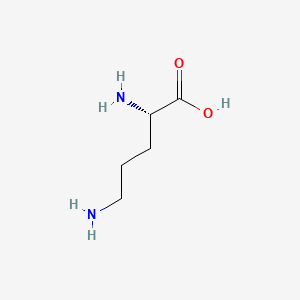 |
0.815 | D0F5DO | 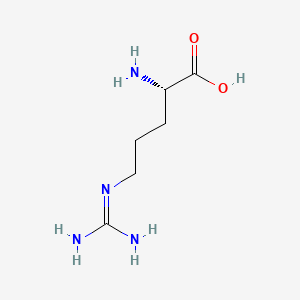 |
0.500 | ||
| ENC000550 | 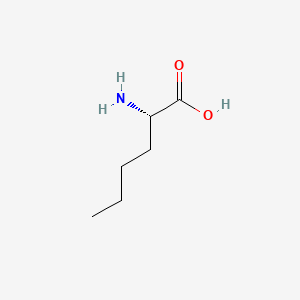 |
0.581 | D0FD0H | 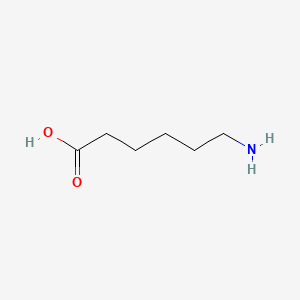 |
0.471 | ||
| ENC000795 |  |
0.500 | D01JIA | 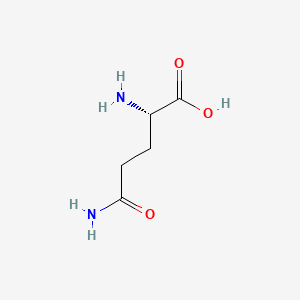 |
0.457 | ||
| ENC000142 | 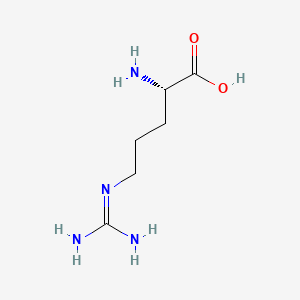 |
0.500 | D01OPV | 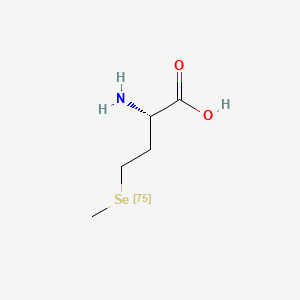 |
0.441 | ||
| ENC000539 | 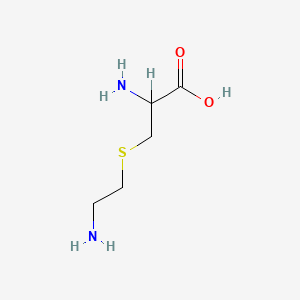 |
0.486 | D00ENY | 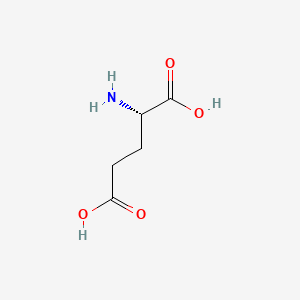 |
0.417 | ||
| ENC000760 | 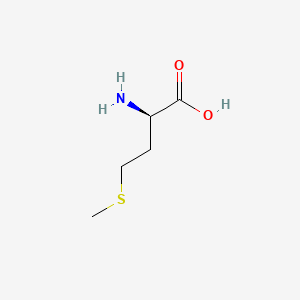 |
0.441 | D03CHT | 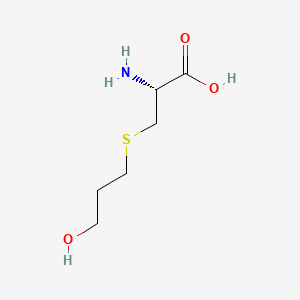 |
0.410 | ||
| ENC000937 | 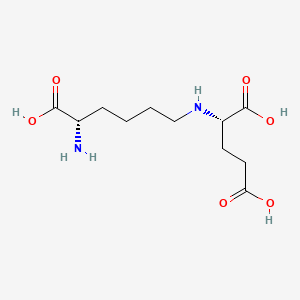 |
0.382 | D02UDJ | 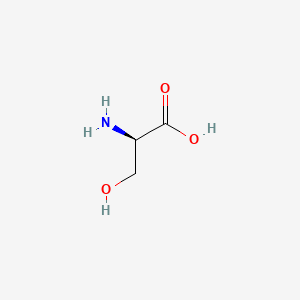 |
0.387 | ||
| ENC000306 | 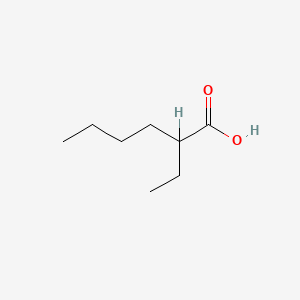 |
0.333 | D0P0QK | 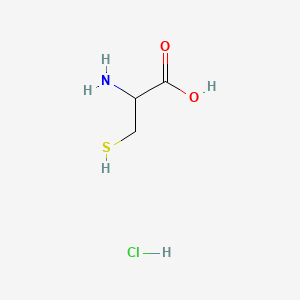 |
0.375 | ||
| ENC001215 | 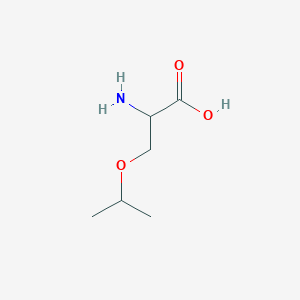 |
0.308 | D0X7JR | 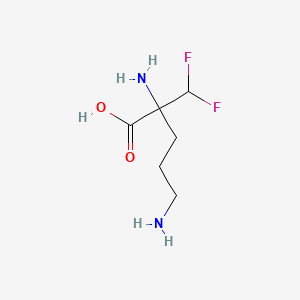 |
0.366 | ||
| ENC000315 | 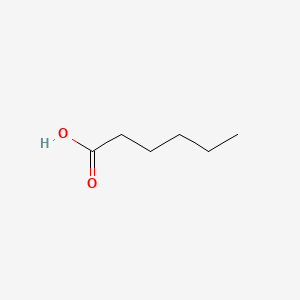 |
0.306 | D00DEF |  |
0.362 | ||