NPs Basic Information
|
Name |
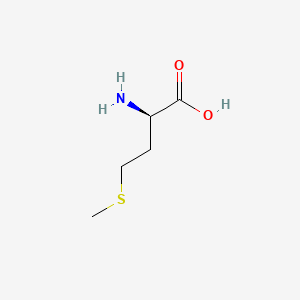
D-Methionine
|
| Molecular Formula | C5H11NO2S | |
| IUPAC Name* |
(2R)-2-amino-4-methylsulfanylbutanoic acid
|
|
| SMILES |
CSCC[C@H](C(=O)O)N
|
|
| InChI |
InChI=1S/C5H11NO2S/c1-9-3-2-4(6)5(7)8/h4H,2-3,6H2,1H3,(H,7,8)/t4-/m1/s1
|
|
| InChIKey |
FFEARJCKVFRZRR-SCSAIBSYSA-N
|
|
| Synonyms |
D-Methionine; 348-67-4; H-D-Met-OH; (R)-Methionine; methionine; D-Methionin; D-Metionien; METHIONINE, D-; (R)-2-amino-4-(methylthio)butanoic acid; (2R)-2-amino-4-(methylsulfanyl)butanoic acid; MRx-1024; Methionine d-form; (2R)-2-amino-4-methylsulfanylbutanoic acid; R-Methionine; CHEBI:16867; D-2-Amino-4-(methylthio)butyric acid; CHEMBL1234268; MED; 868496F25R; D-2-Amino-4-(methylthio)butanoic acid; MFCD00002622; Methionine D-; (R)-2-Amino-4-(methylmercapto)butyric acid; D-Metionien [Australian]; NSC-45689; L-Methionine;; UNII-868496F25R; D-Met; EINECS 206-483-6; NSC 45689; D-Met-OH; bmse000450; D-Methionine;MRX-1024; SCHEMBL126688; S-METHYL-D-HOMOCYSTEINE; METHIONINE D-FORM [MI]; DTXSID90883369; D-Methionine, >=98% (HPLC); ACT04819; ZINC1532766; BDBM50463194; s4494; AKOS015852500; AC-8661; AM81977; CS-7880; DB02893; NCGC00163337-01; AC-12369; DS-15125; HY-13694; (R)-2-Amino-4-methylsulfanyl-butyric acid; D-Methionine, puriss., >=99.0% (NT); DB-038140; M0102; EN300-72154; C00855; 348M674; A822453; J-300192; Q-101531; Q27093862; Z1147215595
|
|
| CAS | 348-67-4 | |
| PubChem CID | 84815 | |
| ChEMBL ID | CHEMBL1234268 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 149.21 | ALogp: | -1.9 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 4 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 88.6 | Aromatic Rings: | 0 |
| Heavy Atoms: | 9 | QED Weighted: | 0.612 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.747 | MDCK Permeability: | 0.00000896 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.026 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.605 | Plasma Protein Binding (PPB): | 11.97% |
| Volume Distribution (VD): | 0.485 | Fu: | 85.90% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.012 | CYP1A2-substrate: | 0.063 |
| CYP2C19-inhibitor: | 0.027 | CYP2C19-substrate: | 0.063 |
| CYP2C9-inhibitor: | 0.009 | CYP2C9-substrate: | 0.319 |
| CYP2D6-inhibitor: | 0.016 | CYP2D6-substrate: | 0.24 |
| CYP3A4-inhibitor: | 0.012 | CYP3A4-substrate: | 0.057 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.051 | Half-life (T1/2): | 0.786 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.015 | Human Hepatotoxicity (H-HT): | 0.08 |
| Drug-inuced Liver Injury (DILI): | 0.018 | AMES Toxicity: | 0.011 |
| Rat Oral Acute Toxicity: | 0.101 | Maximum Recommended Daily Dose: | 0.013 |
| Skin Sensitization: | 0.147 | Carcinogencity: | 0.287 |
| Eye Corrosion: | 0.012 | Eye Irritation: | 0.105 |
| Respiratory Toxicity: | 0.235 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000550 | 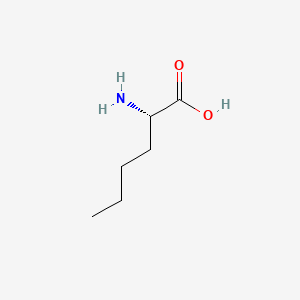 |
0.533 | D01OPV | 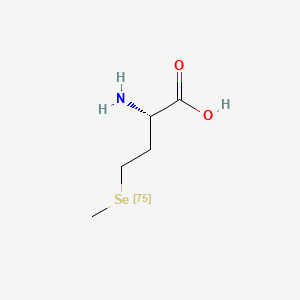 |
0.533 | ||
| ENC000137 | 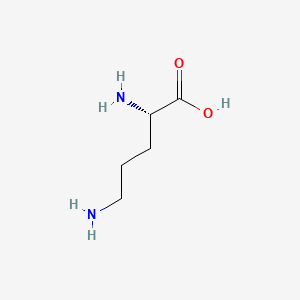 |
0.484 | D01JIA | 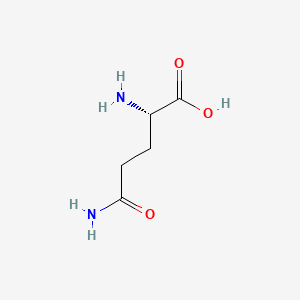 |
0.455 | ||
| ENC000539 | 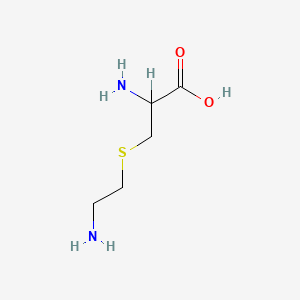 |
0.441 | D00ENY | 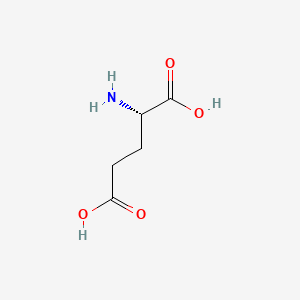 |
0.455 | ||
| ENC000123 | 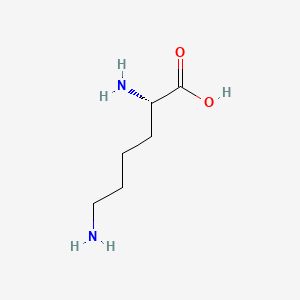 |
0.441 | D02UDJ | 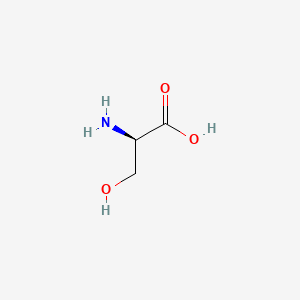 |
0.429 | ||
| ENC000795 |  |
0.417 | D0P0QK | 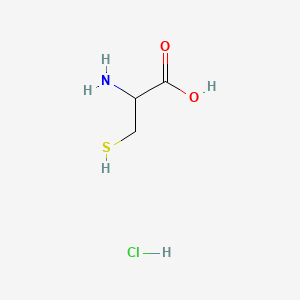 |
0.414 | ||
| ENC000142 | 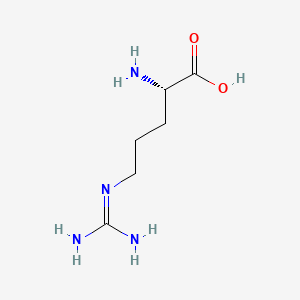 |
0.385 | D03CHT | 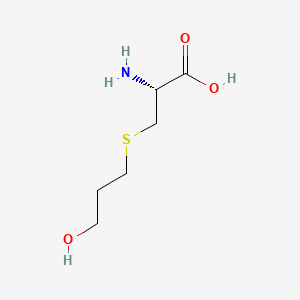 |
0.405 | ||
| ENC001215 | 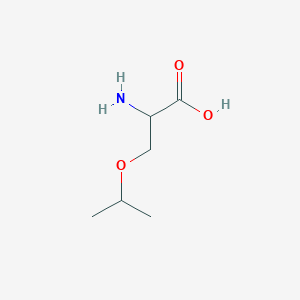 |
0.371 | D0F5DO | 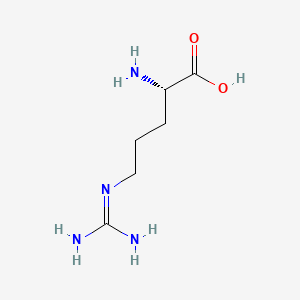 |
0.385 | ||
| ENC000141 | 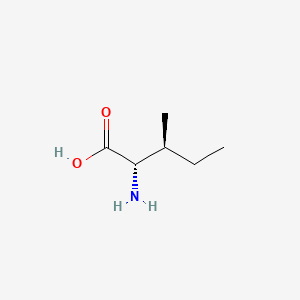 |
0.324 | D09PUL | 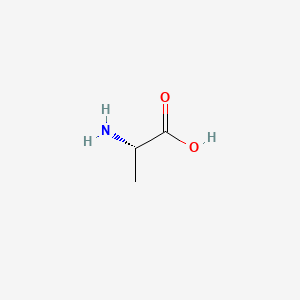 |
0.385 | ||
| ENC000445 | 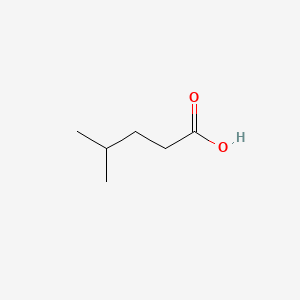 |
0.303 | D0X5SI | 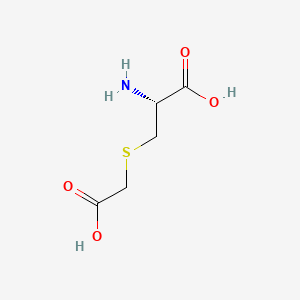 |
0.378 | ||
| ENC000524 | 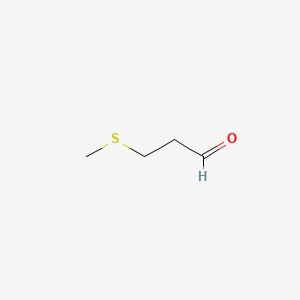 |
0.300 | D0R3QY |  |
0.314 | ||