NPs Basic Information
|
Name |
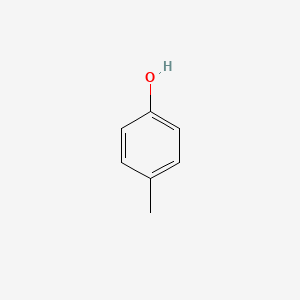
P-Cresol
|
| Molecular Formula | C7H8O | |
| IUPAC Name* |
4-methylphenol
|
|
| SMILES |
CC1=CC=C(C=C1)O
|
|
| InChI |
InChI=1S/C7H8O/c1-6-2-4-7(8)5-3-6/h2-5,8H,1H3
|
|
| InChIKey |
IWDCLRJOBJJRNH-UHFFFAOYSA-N
|
|
| Synonyms |
P-CRESOL; 4-Methylphenol; 106-44-5; 4-Cresol; 4-Hydroxytoluene; Phenol, 4-methyl-; p-Methylphenol; para-Cresol; p-Hydroxytoluene; p-Tolyl alcohol; p-Kresol; p-Oxytoluene; p-Toluol; p-Cresylic acid; 1-Hydroxy-4-methylbenzene; Paracresol; p-Methylhydroxybenzene; 1-Methyl-4-hydroxybenzene; Paramethyl phenol; para-Cresylic acid; 4-methyl phenol; Cresol, para-; Cresol, p-; p-Kresol [German]; Cresol, p-isomer; FEMA No. 2337; CRESOL, PARA; p-Methyl phenol; 4-methyl-phenol; 4-methylphenol (p-cresol); NSC 3696; MFCD00002376; 1MXY2UM8NV; CHEMBL16645; CHEBI:17847; NSC-3696; NCGC00091519-04; TOLUENE,4-HYDROXY (PARA-CRESOL); p-Cresol [UN2076] [Poison, Corrosive]; DSSTox_CID_1869; DSSTox_RID_76375; DSSTox_GSID_21869; 27289-34-5; CAS-106-44-5; CCRIS 647; HSDB 1814; EINECS 203-398-6; UNII-1MXY2UM8NV; para cresol; p-Cresylate; p-methyl-phenol; AI3-00150; 4 -methylphenol; Cresol,p-; para-cresyl alcohol; Phenol, 4-methyI; p-Cresol, 99%; Spectrum_000850; p-Cresol, High Purity; 4-Methyl Phenol 99%; P-CRESOL [FHFI]; P-CRESOL [HSDB]; P-CRESOL [INCI]; Spectrum2_000765; Spectrum4_001740; Spectrum5_000540; P-CRESOL [MI]; SCHEMBL375; bmse000458; EC 203-398-6; DSSTox_RID_77380; DSSTox_RID_77554; NCIOpen2_001516; WLN: QR D1; DSSTox_GSID_24364; DSSTox_GSID_24858; KBioGR_002160; KBioSS_001330; 1-Hydroxyl 4-Methyl Benzene; p-Cresol, analytical standard; BIDD:ER0010; DivK1c_000381; SPECTRUM1500209; p-Cresol, >=99%, FG; SPBio_000810; SCHEMBL7812506; SGCUT00097; DTXSID7021869; HMS501D03; KBio1_000381; KBio2_001330; KBio2_003898; KBio2_006466; NSC3696; PARACRESOL [USP IMPURITY]; NINDS_000381; HMS1920A16; HMS2091I04; Pharmakon1600-01500209; ZINC897142; p-Cresol, for synthesis, 98.0%; 4-Methylphenol, analytical standard; NSC95259; to_000033; Tox21_113240; Tox21_113445; Tox21_200402; Tox21_201115; Tox21_300029; BDBM50008543; CCG-38990; NSC-95259; NSC756709; STL183323; AKOS000119005; Tox21_113445_1; DB01688; IDI1_000381; NCGC00013272-01; NCGC00091519-01; NCGC00091519-02; NCGC00091519-03; NCGC00091519-05; NCGC00091519-06; NCGC00091519-07; NCGC00091519-09; NCGC00253980-01; NCGC00257956-01; NCGC00258667-01; 4-Methylphenol 10 microg/mL in Methanol; PS-11958; CAS-1319-77-3; p-Cresol, JIS special grade, >=99.0%; SBI-0051322.P003; METACRESOL IMPURITY C [EP IMPURITY]; FT-0660000; 4-Methylphenol 100 microg/mL in Cyclohexane; EN300-19427; p-Cresol, puriss. p.a., >=99.0% (GC); AMYLMETACRESOL IMPURITY D [EP IMPURITY]; C01468; AB00051955_02; Q312251; SR-05000002037; J-001591; J-515803; SR-05000002037-1; F1908-0066; Z104473818; 2876-02-0
|
|
| CAS | 106-44-5 | |
| PubChem CID | 2879 | |
| ChEMBL ID | CHEMBL16645 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 108.14 | ALogp: | 1.9 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.54 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.341 | MDCK Permeability: | 0.00002350 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.099 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.08 |
| 30% Bioavailability (F30%): | 0.046 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.226 | Plasma Protein Binding (PPB): | 68.24% |
| Volume Distribution (VD): | 1.986 | Fu: | 24.05% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.908 | CYP1A2-substrate: | 0.906 |
| CYP2C19-inhibitor: | 0.653 | CYP2C19-substrate: | 0.647 |
| CYP2C9-inhibitor: | 0.152 | CYP2C9-substrate: | 0.878 |
| CYP2D6-inhibitor: | 0.349 | CYP2D6-substrate: | 0.885 |
| CYP3A4-inhibitor: | 0.053 | CYP3A4-substrate: | 0.346 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 15.598 | Half-life (T1/2): | 0.894 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.027 | Human Hepatotoxicity (H-HT): | 0.038 |
| Drug-inuced Liver Injury (DILI): | 0.045 | AMES Toxicity: | 0.048 |
| Rat Oral Acute Toxicity: | 0.748 | Maximum Recommended Daily Dose: | 0.048 |
| Skin Sensitization: | 0.689 | Carcinogencity: | 0.615 |
| Eye Corrosion: | 0.984 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.42 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001021 | 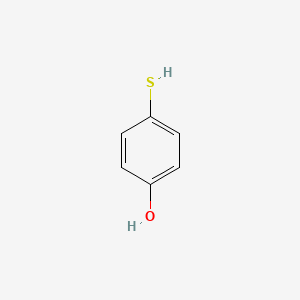 |
0.571 | D03UOT | 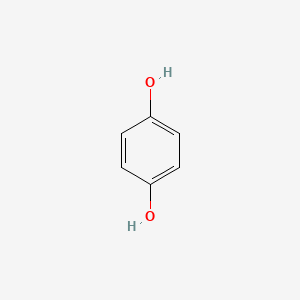 |
0.571 | ||
| ENC000233 | 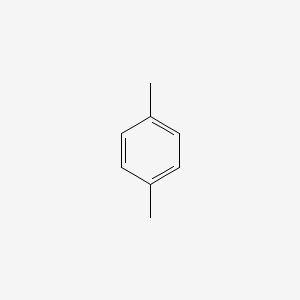 |
0.571 | D0U5QK | 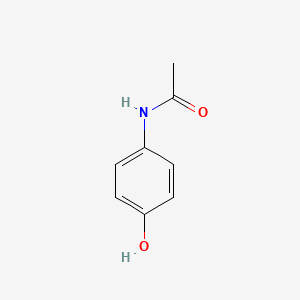 |
0.486 | ||
| ENC000318 | 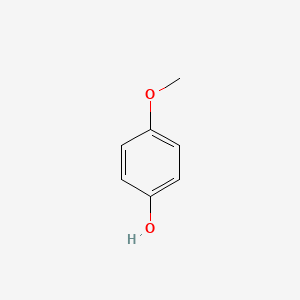 |
0.567 | D0W1RY | 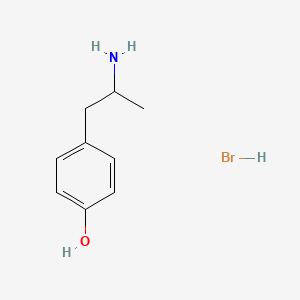 |
0.472 | ||
| ENC000200 | 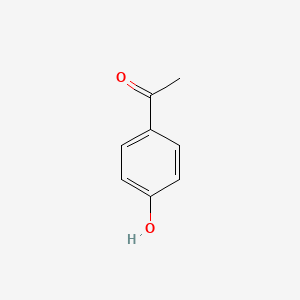 |
0.531 | D02WAB | 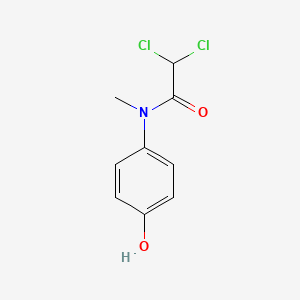 |
0.405 | ||
| ENC000005 | 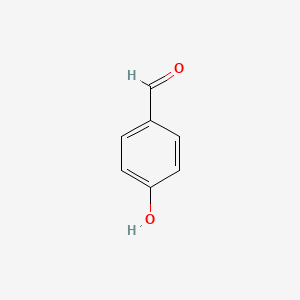 |
0.516 | D0B3QM | 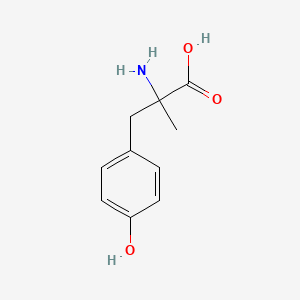 |
0.405 | ||
| ENC000221 | 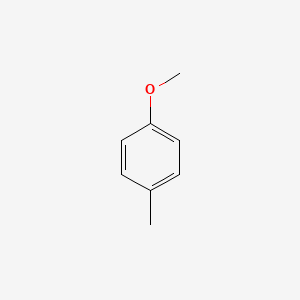 |
0.516 | D01CRB | 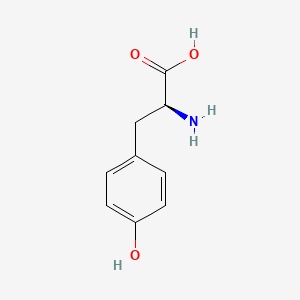 |
0.390 | ||
| ENC000740 | 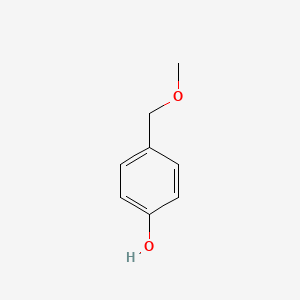 |
0.515 | D06GIP | 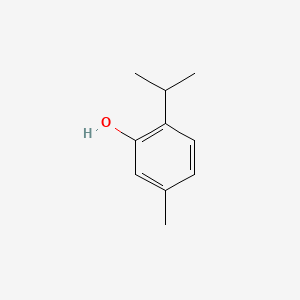 |
0.342 | ||
| ENC000072 | 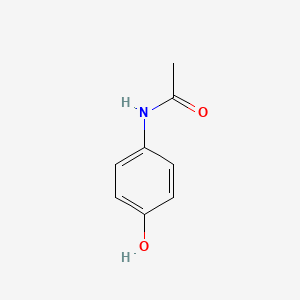 |
0.486 | D0S2BV | 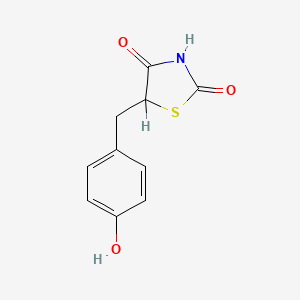 |
0.333 | ||
| ENC000195 | 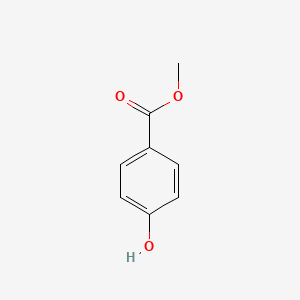 |
0.486 | D0H6TP | 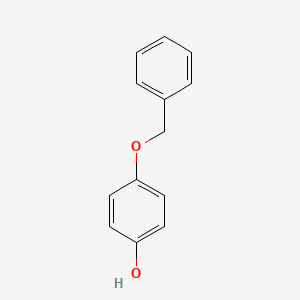 |
0.320 | ||
| ENC000199 | 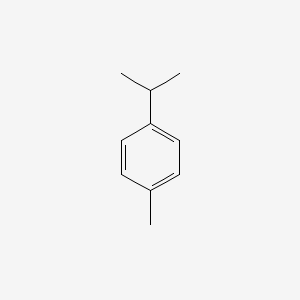 |
0.485 | D0Y7PG | 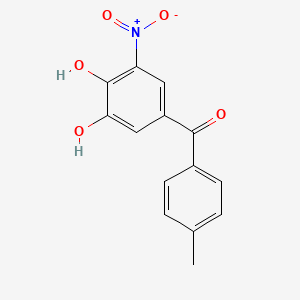 |
0.310 | ||