NPs Basic Information
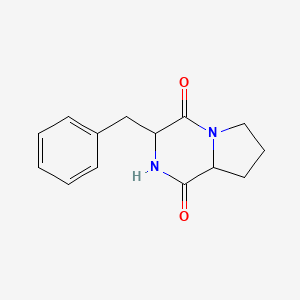
|
Name |
Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)-
|
| Molecular Formula | C14H16N2O2 | |
| IUPAC Name* |
3-benzyl-2,3,6,7,8,8a-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione
|
|
| SMILES |
C1CC2C(=O)NC(C(=O)N2C1)CC3=CC=CC=C3
|
|
| InChI |
InChI=1S/C14H16N2O2/c17-13-12-7-4-8-16(12)14(18)11(15-13)9-10-5-2-1-3-6-10/h1-3,5-6,11-12H,4,7-9H2,(H,15,17)
|
|
| InChIKey |
QZBUWPVZSXDWSB-UHFFFAOYSA-N
|
|
| Synonyms |
14705-60-3; Cyclo(Phe-Pro); Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)-; Cyclo(D-phenylalanyl-L-prolyl); Cyclo(phenylalanylprolyl); Cyclo(L-prolyl-D-phenylalanyl); A 64863; 3-benzyl-2,3,6,7,8,8a-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione; Pyrrolo(2,1-f)pyrazine-1,4-dione, 2,3,6,7,8,8a-hexahydro-3-(phenylmethyl)-; NSC646119; Phenylalanyl-prolyl diketopiperazine; PYRROLO[1,2-A]PYRAZINE-1,4-DIONE,HEXAHYDRO-3-(PHENYLMETHYL)-; 3-benzyl-octahydropyrrolo[1,2-a]pyrazine-1,4-dione; 3-Benzylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione; 26488-24-4; Phe-pro-diketopiperazine; MFCD00038604; L-Phe-D-Pro lactam; NSC 255998; (3S-trans)-3-Benzylhexahydropyrrolo(1,2-a)pyrazine-1,4-dione; SCHEMBL173749; cis-Cyclo(L-phenyl,L-prolyl); Cyclo-Phe-Pro-diketopiperazine; MEGxm0_000490; CHEMBL1990465; ACon0_001420; DTXSID30933019; A-64863Cyclo(phenylalanylprolyl); BCP32901; HY-P1934; EINECS 223-047-0; NSC255998; 3-(Phenylmethyl)-2,3,6,7,8,8a-hexahydropyrrolo(2,1-f)pyrazine-1,4-dione; AKOS015999981; NSC-255998; NSC-646119; NCGC00380633-02; AS-71116; NCI60_015870; SY045657; DB-049055; CS-0064705; FT-0634981; D92888; Cyclo(-Phe-Pro);Cyclo-L-phenylalanyl-L-proline; A929346; 3-Benzylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione #; Pyrrolo[1,4-dione, hexahydro-3-(phenylmethyl)-, (3R-cis)-; 3-Benzyl-1-hydroxy-6,7,8,8a-tetrahydropyrrolo[1,2-a]pyrazin-4(3H)-one; Pyrrolo(1,2-a)pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)-, (3R-cis)-; NCGC00380633-02_C14H16N2O2_Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)-
|
|
| CAS | 3705-26-8 | |
| PubChem CID | 99895 | |
| ChEMBL ID | CHEMBL1990465 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 244.29 | ALogp: | 1.4 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 49.4 | Aromatic Rings: | 3 |
| Heavy Atoms: | 18 | QED Weighted: | 0.847 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.694 | MDCK Permeability: | 0.00005270 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.018 | 20% Bioavailability (F20%): | 0.065 |
| 30% Bioavailability (F30%): | 0.05 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.701 | Plasma Protein Binding (PPB): | 47.96% |
| Volume Distribution (VD): | 0.577 | Fu: | 44.41% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.037 | CYP1A2-substrate: | 0.121 |
| CYP2C19-inhibitor: | 0.338 | CYP2C19-substrate: | 0.496 |
| CYP2C9-inhibitor: | 0.086 | CYP2C9-substrate: | 0.75 |
| CYP2D6-inhibitor: | 0.011 | CYP2D6-substrate: | 0.446 |
| CYP3A4-inhibitor: | 0.174 | CYP3A4-substrate: | 0.283 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.155 | Half-life (T1/2): | 0.74 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.751 |
| Drug-inuced Liver Injury (DILI): | 0.187 | AMES Toxicity: | 0.016 |
| Rat Oral Acute Toxicity: | 0.726 | Maximum Recommended Daily Dose: | 0.316 |
| Skin Sensitization: | 0.214 | Carcinogencity: | 0.133 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.018 |
| Respiratory Toxicity: | 0.041 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005971 | 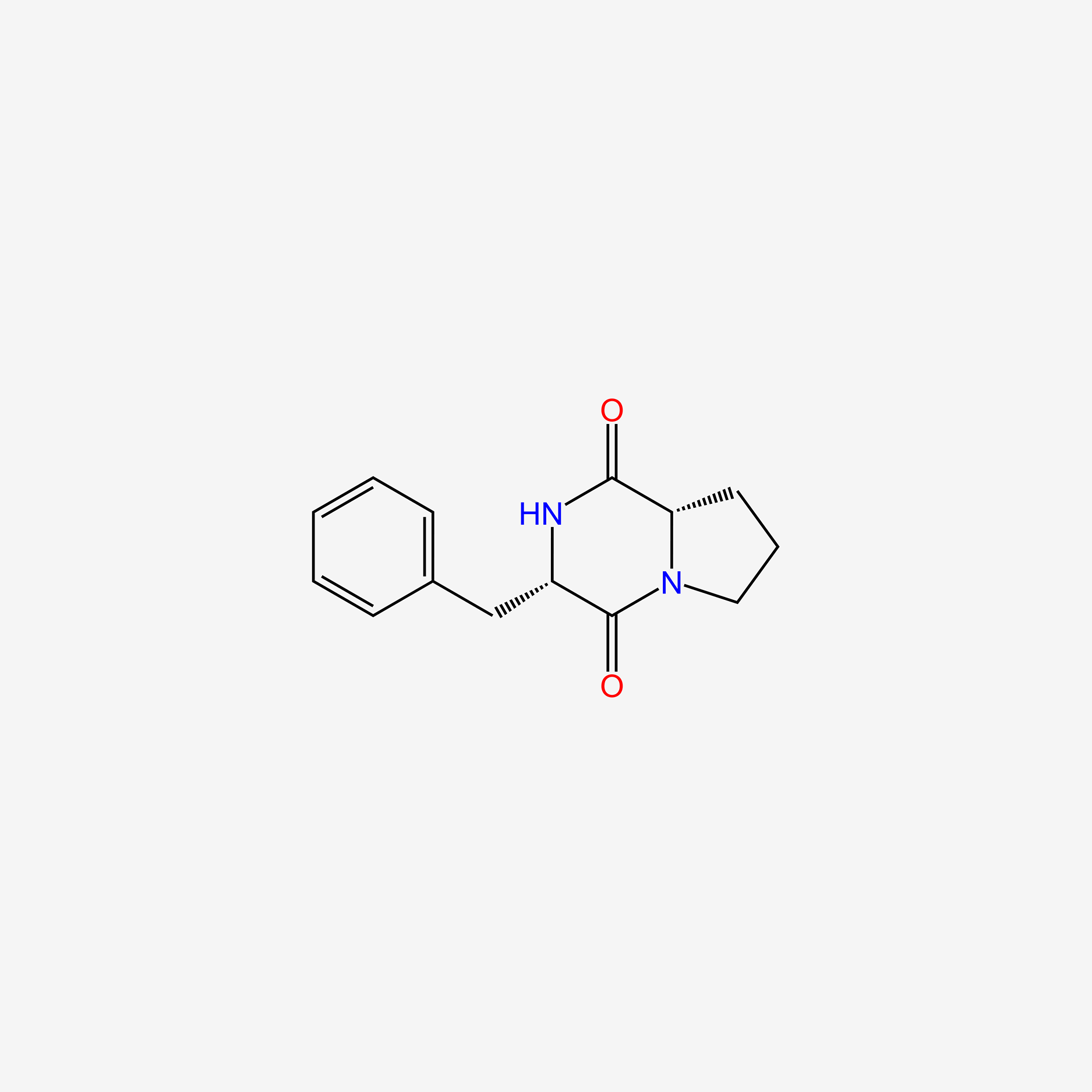 |
1.000 | D05EPM | 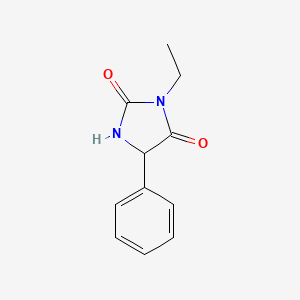 |
0.382 | ||
| ENC005484 | 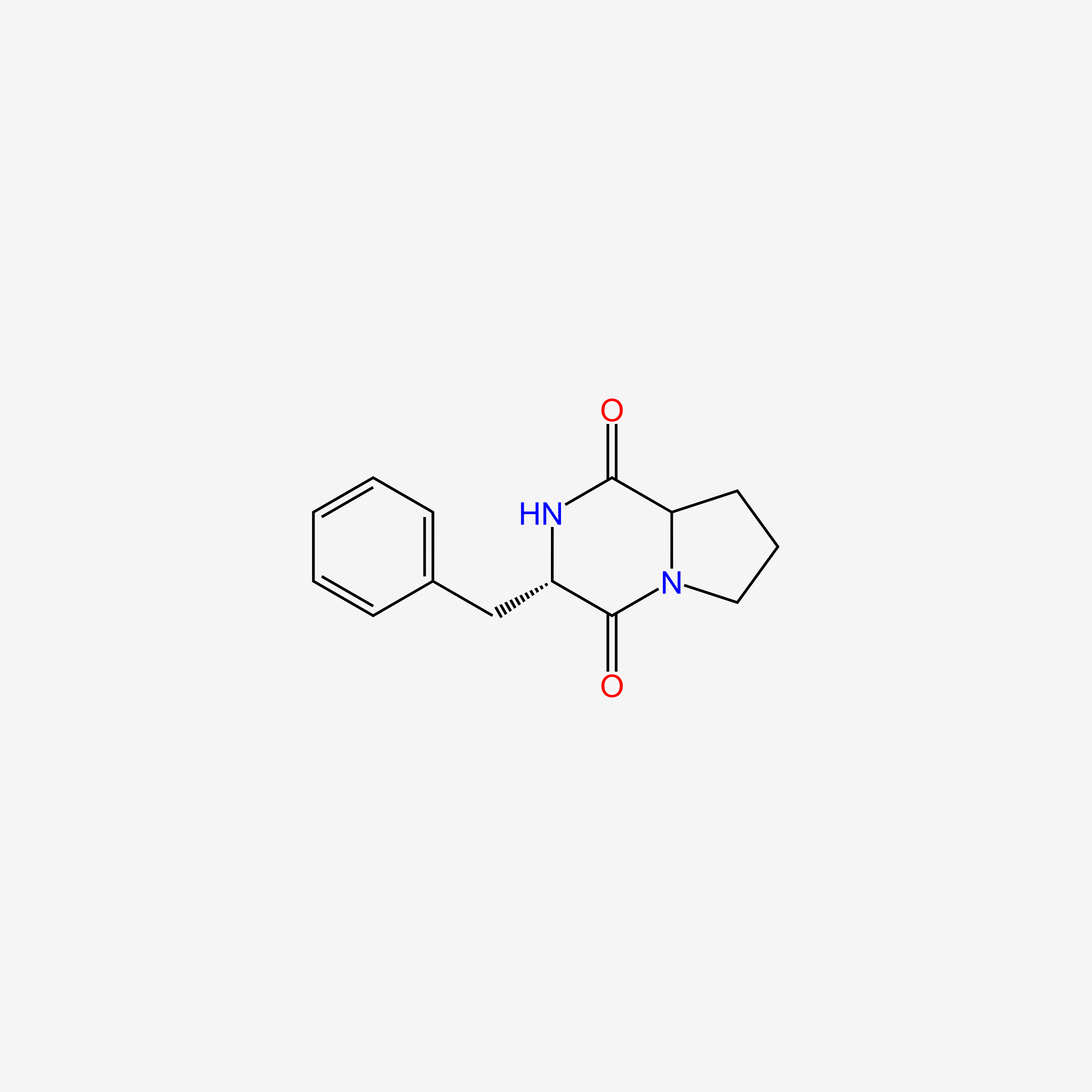 |
1.000 | D06BYV | 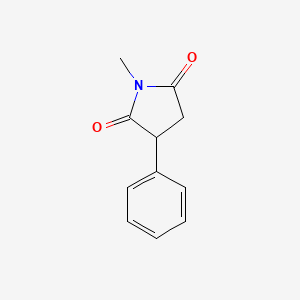 |
0.358 | ||
| ENC005206 | 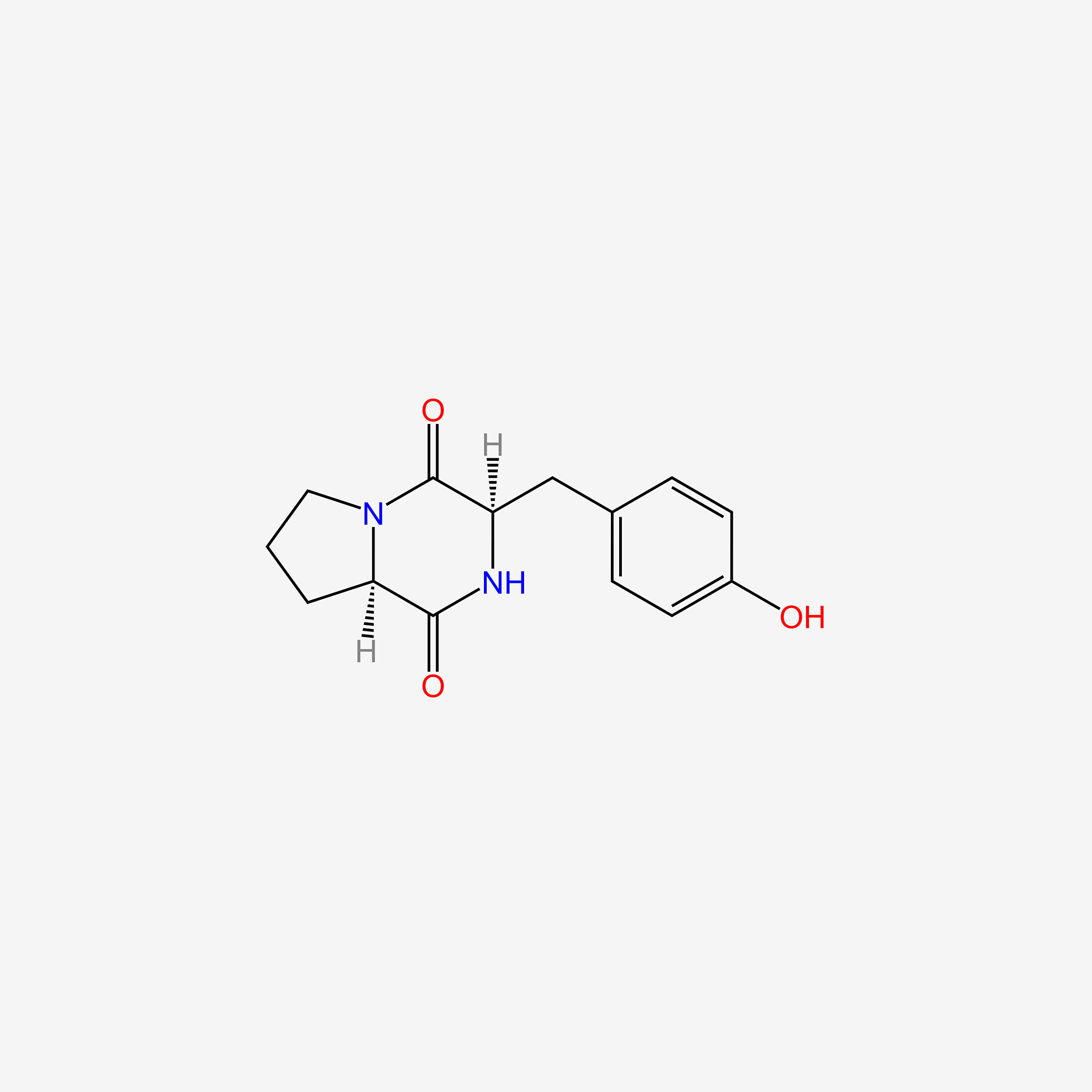 |
0.683 | D03RZV | 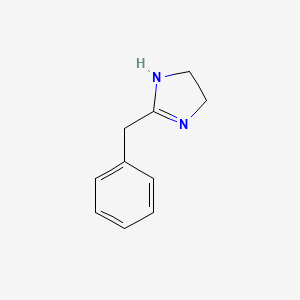 |
0.333 | ||
| ENC005847 | 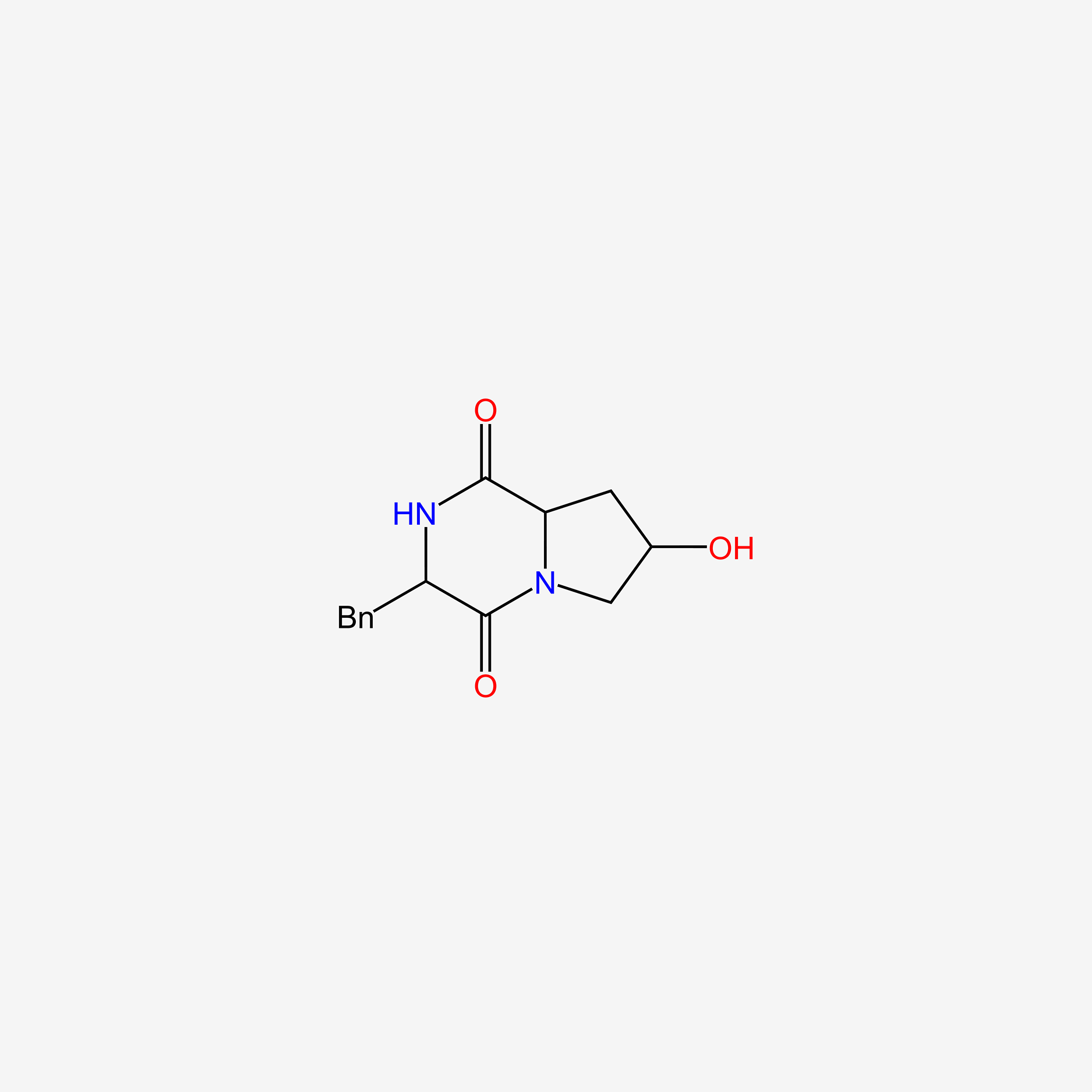 |
0.683 | D0Z9NZ | 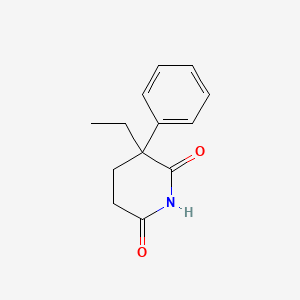 |
0.329 | ||
| ENC005092 |  |
0.683 | D04GKO | 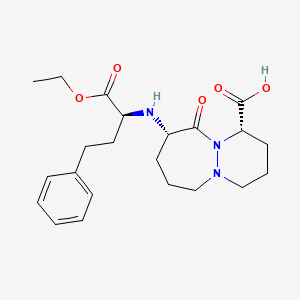 |
0.317 | ||
| ENC005408 | 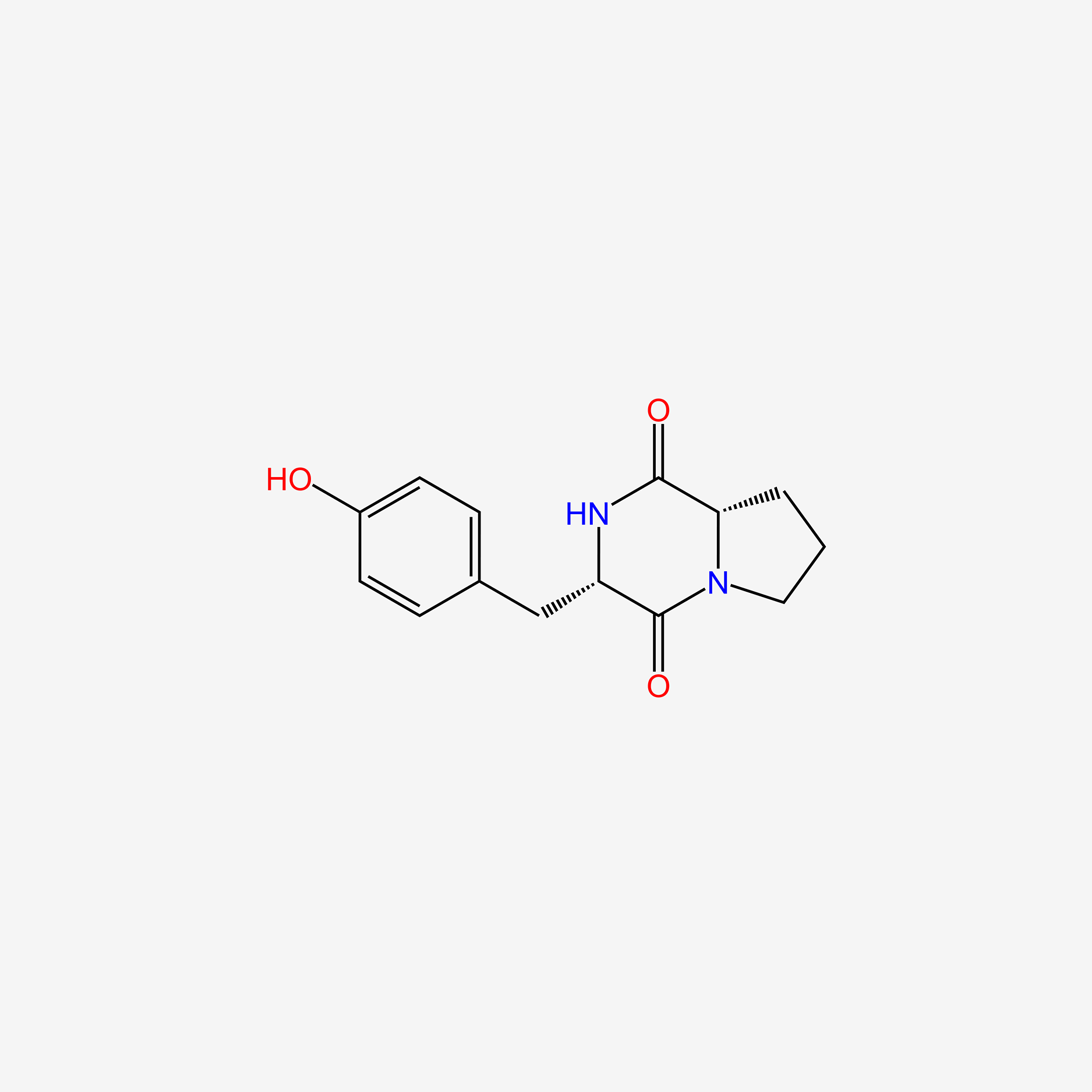 |
0.683 | D05OIS | 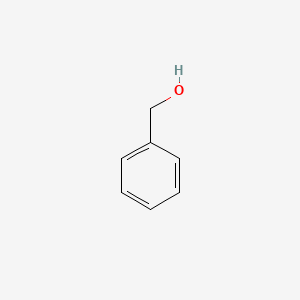 |
0.316 | ||
| ENC000867 | 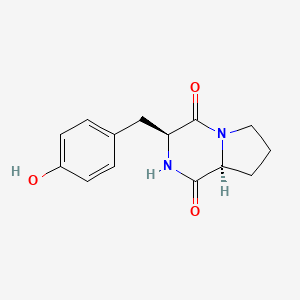 |
0.683 | D0I0DL | 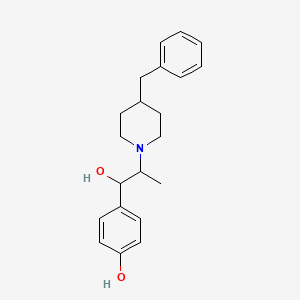 |
0.315 | ||
| ENC000975 | 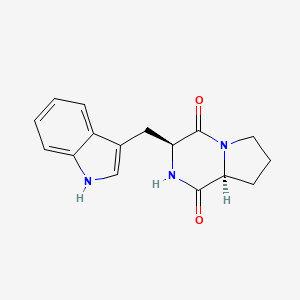 |
0.662 | D0N5HJ | 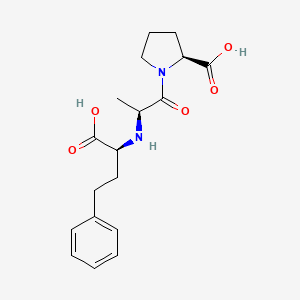 |
0.315 | ||
| ENC003217 | 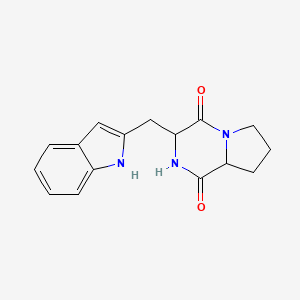 |
0.662 | D07RGW | 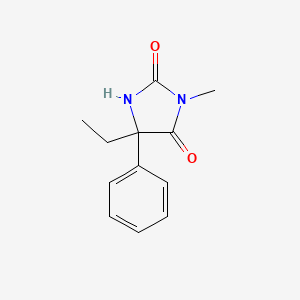 |
0.315 | ||
| ENC005969 |  |
0.606 | D02DMQ | 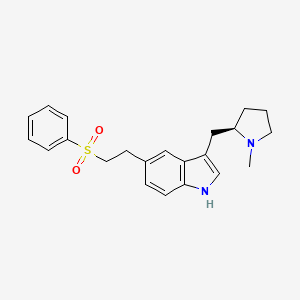 |
0.313 | ||