NPs Basic Information
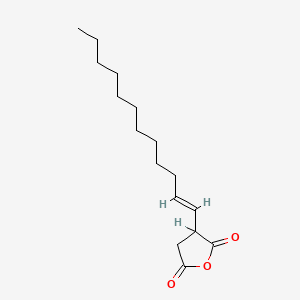
|
Name |
Dodecenylsuccinic anhydride
|
| Molecular Formula | C16H26O3 | |
| IUPAC Name* |
3-[(E)-dodec-1-enyl]oxolane-2,5-dione
|
|
| SMILES |
CCCCCCCCCC/C=C/C1CC(=O)OC1=O
|
|
| InChI |
InChI=1S/C16H26O3/c1-2-3-4-5-6-7-8-9-10-11-12-14-13-15(17)19-16(14)18/h11-12,14H,2-10,13H2,1H3/b12-11+
|
|
| InChIKey |
WVRNUXJQQFPNMN-VAWYXSNFSA-N
|
|
| Synonyms |
DODECENYLSUCCINIC ANHYDRIDE; 25377-73-5; 2,5-Furandione, 3-(dodecenyl)dihydro-; DDSA; 3-[(E)-dodec-1-enyl]oxolane-2,5-dione; ALQ23WB445; 3-[(1E)-dodec-1-en-1-yl]oxolane-2,5-dione; n-Dodecenylsuccinic anhydride; 1-dodecenylsuccinic anhydride; 119295-58-8; 2,5-Furandione, 3-(1-dodecenyl)dihydro-, (E)-; 19532-92-4; Rikacid DDSA; K 12 (anhydride); Dodecenyl succinic anhydride; CCRIS 698; 2-(Dodecyl)succinic anhydride; Succinic anhydride, dodecenyl-; Dodecenylsuccinic acid anhydride; HSDB 4374; EINECS 246-917-1; UNII-ALQ23WB445; SCHEMBL107779; DTXSID80893538; 2-(1-Dodecenyl)succinic anhydride; AKOS015912885; NCGC00164187-02; AS-81577; 2,5-Furandione, 3-(dodecen-1-yl)dihydro-; (E)-3-(dodec-1-enyl)dihydrofuran-2,5-dione; 3-[(1E)-1-Dodecen-1-yl]dihydro-2,5-furandione; W-109191
|
|
| CAS | 25377-73-5 | |
| PubChem CID | 6433892 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 266.38 | ALogp: | 5.4 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 10 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 43.4 | Aromatic Rings: | 1 |
| Heavy Atoms: | 19 | QED Weighted: | 0.248 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.834 | MDCK Permeability: | 0.00002620 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.009 |
| Human Intestinal Absorption (HIA): | 0.014 | 20% Bioavailability (F20%): | 0.96 |
| 30% Bioavailability (F30%): | 0.974 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.218 | Plasma Protein Binding (PPB): | 99.80% |
| Volume Distribution (VD): | 3.79 | Fu: | 1.78% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.686 | CYP1A2-substrate: | 0.38 |
| CYP2C19-inhibitor: | 0.354 | CYP2C19-substrate: | 0.105 |
| CYP2C9-inhibitor: | 0.423 | CYP2C9-substrate: | 0.97 |
| CYP2D6-inhibitor: | 0.17 | CYP2D6-substrate: | 0.688 |
| CYP3A4-inhibitor: | 0.082 | CYP3A4-substrate: | 0.051 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.74 | Half-life (T1/2): | 0.682 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.197 |
| Drug-inuced Liver Injury (DILI): | 0.03 | AMES Toxicity: | 0.025 |
| Rat Oral Acute Toxicity: | 0.145 | Maximum Recommended Daily Dose: | 0.161 |
| Skin Sensitization: | 0.947 | Carcinogencity: | 0.545 |
| Eye Corrosion: | 0.877 | Eye Irritation: | 0.92 |
| Respiratory Toxicity: | 0.898 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001656 | 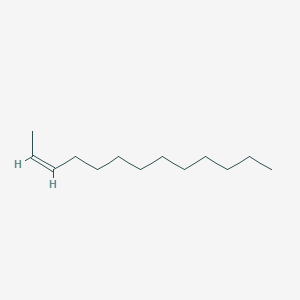 |
0.542 | D03ZJE | 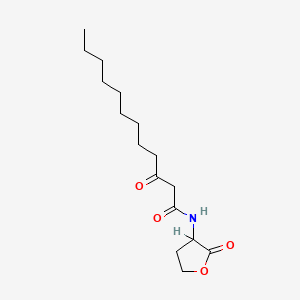 |
0.395 | ||
| ENC000275 | 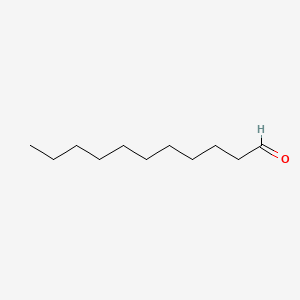 |
0.517 | D05ATI |  |
0.384 | ||
| ENC001644 | 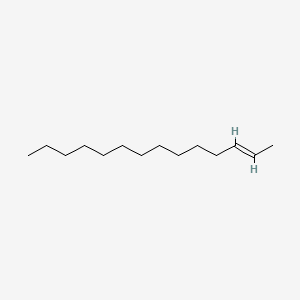 |
0.516 | D0O1PH |  |
0.379 | ||
| ENC000277 | 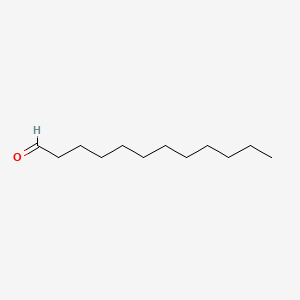 |
0.492 | D0I4DQ | 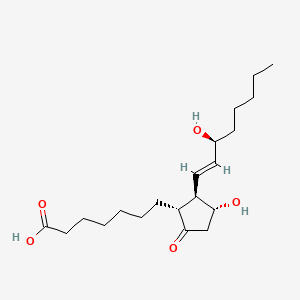 |
0.352 | ||
| ENC001655 | 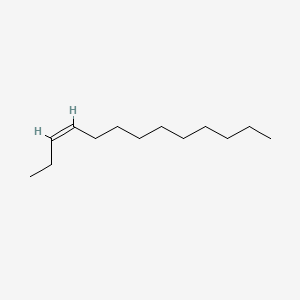 |
0.492 | D0Z5SM |  |
0.350 | ||
| ENC000273 | 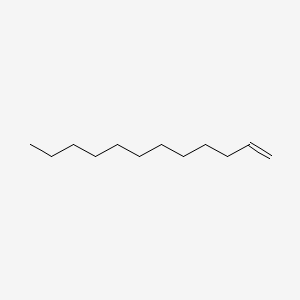 |
0.492 | D0Y8DP | 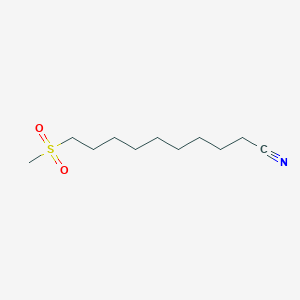 |
0.338 | ||
| ENC001588 | 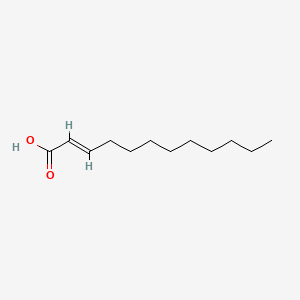 |
0.476 | D0O1TC | 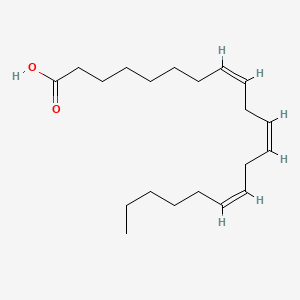 |
0.330 | ||
| ENC001685 |  |
0.471 | D0Z5BC |  |
0.324 | ||
| ENC000510 |  |
0.468 | D09ANG |  |
0.320 | ||
| ENC000267 | 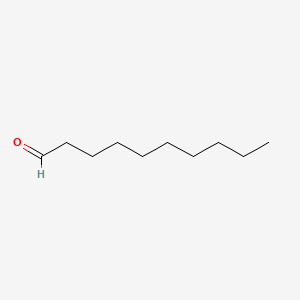 |
0.466 | D0XN8C | 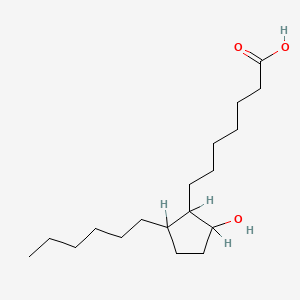 |
0.314 | ||