NPs Basic Information

|
Name |
Piperlongumine
|
| Molecular Formula | C17H19NO5 | |
| IUPAC Name* |
1-[(E)-3-(3,4,5-trimethoxyphenyl)prop-2-enoyl]-2,3-dihydropyridin-6-one
|
|
| SMILES |
COC1=CC(=CC(=C1OC)OC)/C=C/C(=O)N2CCC=CC2=O
|
|
| InChI |
InChI=1S/C17H19NO5/c1-21-13-10-12(11-14(22-2)17(13)23-3)7-8-16(20)18-9-5-4-6-15(18)19/h4,6-8,10-11H,5,9H2,1-3H3/b8-7+
|
|
| InChIKey |
VABYUUZNAVQNPG-BQYQJAHWSA-N
|
|
| Synonyms |
Piperlongumine; Piplartine; 20069-09-4; Piperlongumin; (E)-1-(3-(3,4,5-trimethoxyphenyl)acryloyl)-5,6-dihydropyridin-2(1H)-one; PPLGM; CHEBI:8241; SGD66V4SVJ; 1-[(2E)-3-(3,4,5-trimethoxyphenyl)prop-2-enoyl]-1,2,5,6-tetrahydropyridin-2-one; 1-[(E)-3-(3,4,5-trimethoxyphenyl)prop-2-enoyl]-2,3-dihydropyridin-6-one; BRD2293; BRD-2293; 2(1H)-Pyridinone, 5,6-dihydro-1-[(2E)-1-oxo-3-(3,4,5-trimethoxyphenyl)-2-propen-1-yl]-; UNII-SGD66V4SVJ; (E)-1-[3-(3,4,5-Trimethoxyphenyl)acryloyl]-5,6-dihydropyridin-2(1H)-one; Piplartine;PPLGM; Prestwick_399; MFCD00075706; FERROUSFLUOBORATE; ST079382; Prestwick2_000604; Prestwick3_000604; Piperlongumine; Piplartine; PIPERLONGUMINE [MI]; 5,6-Dihydro-1-[(2E)-1-oxo-3-(3,4,5-trimethoxyphenyl)-2-propen-1-yl]-2(1H)-pyridinone; BSPBio_000508; PIPERLONGUMINE [INCI]; MLS002153903; SCHEMBL173092; SPECTRUM1505135; BPBio1_000560; CHEMBL465843; SCHEMBL2465593; 1-[3-(3,4,5-Trimethoxy-phenyl)-acryloyl]-5,6-dihydro-1H-pyridin-2-one; ACon1_001541; CHEBI:92424; DTXSID801029762; HMS1569J10; HMS2096J10; HMS2234K24; Piperlongumine, >=97% (HPLC); ZINC899053; BCP13030; EX-A2925; HY-N2329; BDBM50462013; NSC794671; s7551; AKOS024284776; CCG-214375; NSC-794671; 2(1H)-Pyridinone, 5,6-dihydro-1-(1-oxo-3-(3,4,5-trimethoxyphenyl)-2-propenyl)-, (E)-; NCGC00096028-01; NCGC00096028-02; NCGC00096028-03; NCGC00096028-04; NCGC00096028-14; AC-32683; AS-74140; BP-25401; LS-14579; SMR001233252; CS-0021113; P2361; A14124; EN300-7424497; A920539; SR-01000841248; A1-00162; J-012992; N-(3,4,5-Trimethoxycinnamoyl)-D3-piperidin-2-one; Q7197361; SR-01000841248-2; BRD-K24132293-001-05-3; BRD-K24132293-001-09-5; BRD-K24132293-001-16-0; 5,6-Dihydro-1-(3,4,5-trimethoxycinnamoyl)-2(1H)-pyridinone; 1-[(2E)-3-(3,4,5-Trimethoxyphenyl)-2-propenoyl]-5,6-dihydro-2(1H)-pyridinone; 1-[(2E)-3-(3,4,5-Trimethoxyphenyl)-2-propenoyl]-5,6-dihydro-2(1H)-pyridinone #; 1-[(2E)-3-(3,4,5-trimethoxyphenyl)prop-2-enoyl]-5,6-dihydropyridin-2(1H)-one; 5,6-Dihydro-1-[1-oxo-3-(3,4,5-trimethoxyphenyl)-2-propenyl]-2(1H)-pyridinone, 9CI; 5,6-dihydro-1-[1-oxo-3-(3,4,5-triMethoxyphenyl)-allyl]-2(1H)-pyridinone; 5,6-Dihydro-1-[1-oxo-3-(3,4,5-trimethoxyphenyl)-trans-2-propenyl]-2(1H)-pyridinone; Prop-2-en-1-one, 3-(3,4,5-trimethoxyphenyl)-1-(2,3-dihydropyridin-6(1H)-one-1-yl)-; (2E)-1-(1,2,5,6-Tetrahydro-2-oxopyridine-1-yl)-3-(3,4,5-trimethoxyphenyl)-2-propene-1-one; 2(1H)-PYRIDINONE, 5,6-DIHYDRO-1-(1-OXO-3-(3,4,5-TRIMETHOXYPHENYL)-2- PROPENYL)-, (E)-
|
|
| CAS | 20069-09-4 | |
| PubChem CID | 637858 | |
| ChEMBL ID | CHEMBL465843 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 317.34 | ALogp: | 2.1 |
| HBD: | 0 | HBA: | 5 |
| Rotatable Bonds: | 5 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 65.1 | Aromatic Rings: | 2 |
| Heavy Atoms: | 23 | QED Weighted: | 0.781 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.6 | MDCK Permeability: | 0.00001440 |
| Pgp-inhibitor: | 0.515 | Pgp-substrate: | 0.225 |
| Human Intestinal Absorption (HIA): | 0.051 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.897 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.985 | Plasma Protein Binding (PPB): | 51.10% |
| Volume Distribution (VD): | 0.421 | Fu: | 21.46% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.82 | CYP1A2-substrate: | 0.964 |
| CYP2C19-inhibitor: | 0.185 | CYP2C19-substrate: | 0.46 |
| CYP2C9-inhibitor: | 0.103 | CYP2C9-substrate: | 0.889 |
| CYP2D6-inhibitor: | 0.3 | CYP2D6-substrate: | 0.337 |
| CYP3A4-inhibitor: | 0.116 | CYP3A4-substrate: | 0.431 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.343 | Half-life (T1/2): | 0.816 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.049 | Human Hepatotoxicity (H-HT): | 0.152 |
| Drug-inuced Liver Injury (DILI): | 0.126 | AMES Toxicity: | 0.04 |
| Rat Oral Acute Toxicity: | 0.01 | Maximum Recommended Daily Dose: | 0.046 |
| Skin Sensitization: | 0.93 | Carcinogencity: | 0.687 |
| Eye Corrosion: | 0.014 | Eye Irritation: | 0.181 |
| Respiratory Toxicity: | 0.026 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005523 |  |
0.684 | D09DHY | 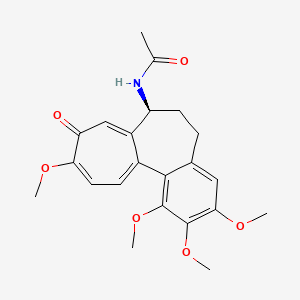 |
0.346 | ||
| ENC001396 | 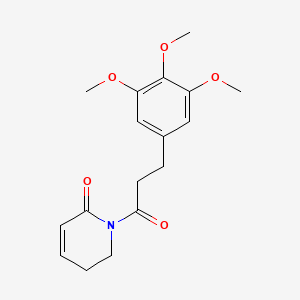 |
0.662 | D0AO5H | 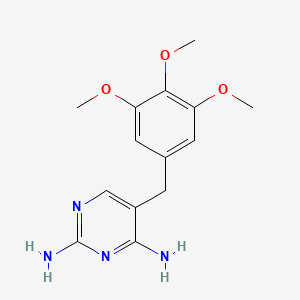 |
0.341 | ||
| ENC000340 | 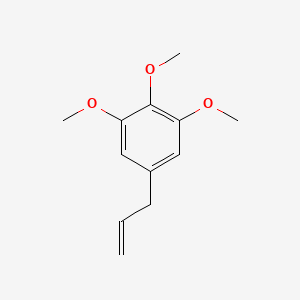 |
0.400 | D0G8NJ | 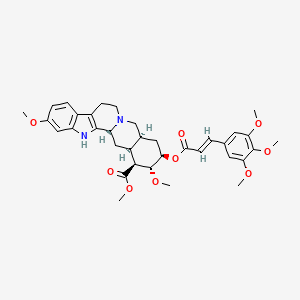 |
0.338 | ||
| ENC001376 | 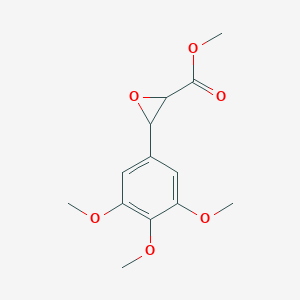 |
0.381 | D0E6OC | 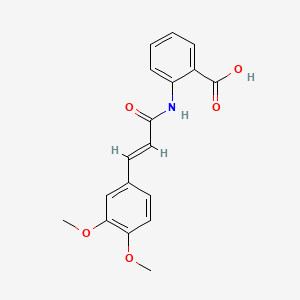 |
0.337 | ||
| ENC005314 | 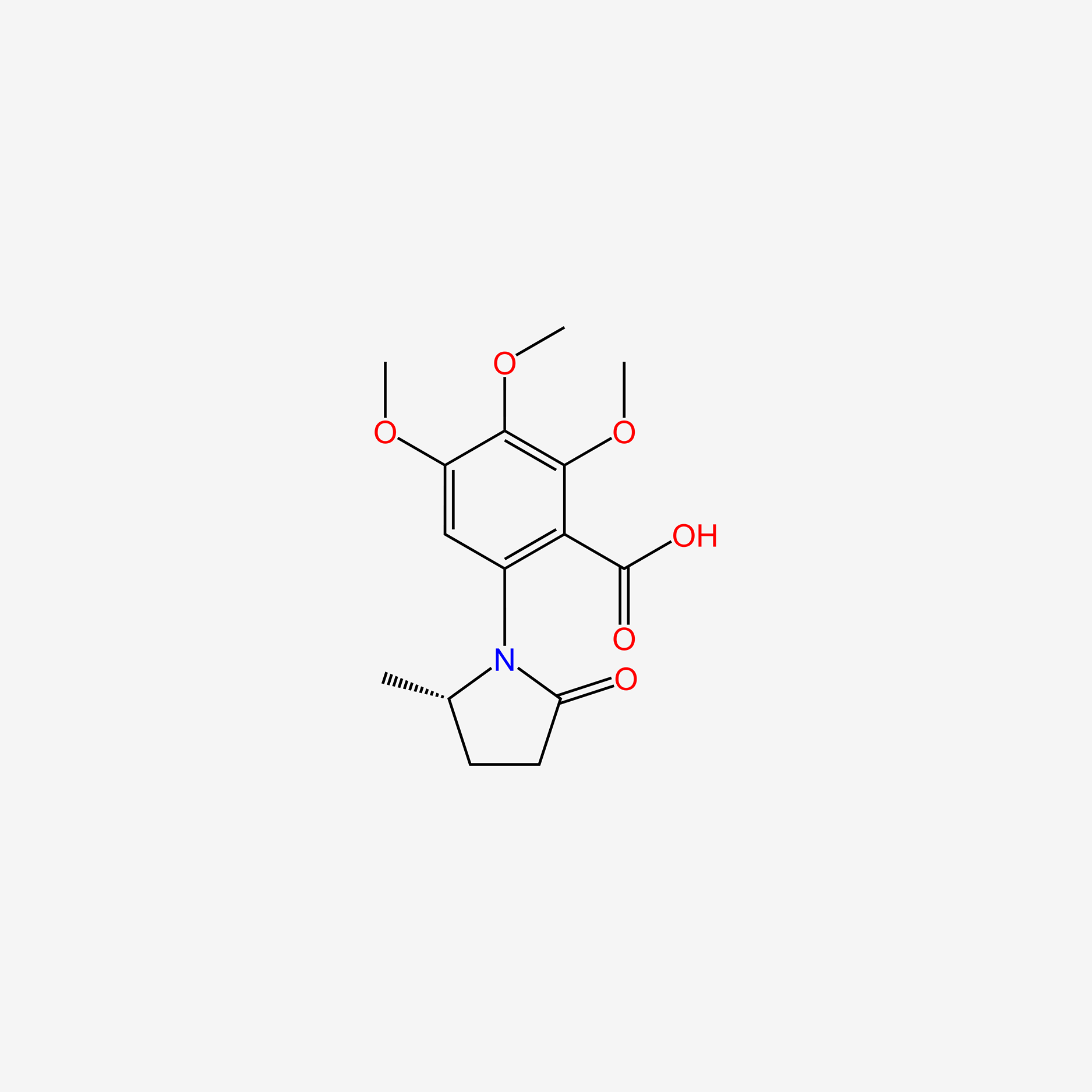 |
0.352 | D0A8FB | 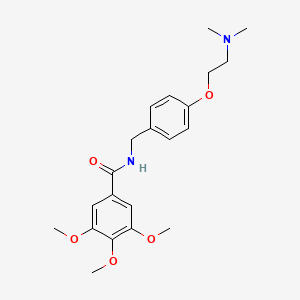 |
0.327 | ||
| ENC001577 | 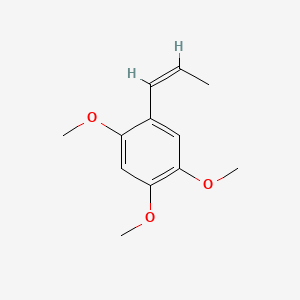 |
0.346 | D02LZB | 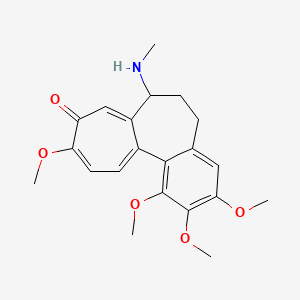 |
0.324 | ||
| ENC001410 | 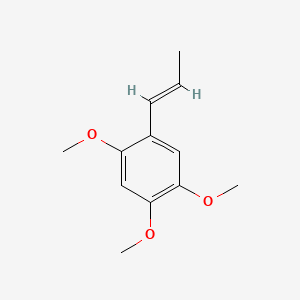 |
0.346 | D0D4HN |  |
0.319 | ||
| ENC001416 |  |
0.340 | D01FFA |  |
0.314 | ||
| ENC000304 |  |
0.338 | D06GCK |  |
0.304 | ||
| ENC001461 | 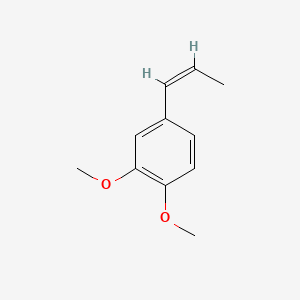 |
0.333 | D0Y7TS | 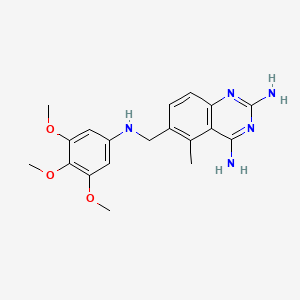 |
0.299 | ||