NPs Basic Information
|
Name |
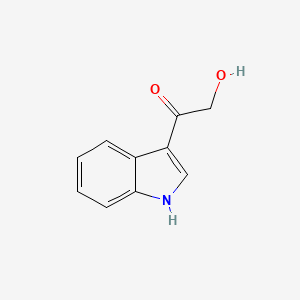
2-Hydroxy-1-(1H-indol-3-yl)ethanone
|
| Molecular Formula | C10H9NO2 | |
| IUPAC Name* |
2-hydroxy-1-(1H-indol-3-yl)ethanone
|
|
| SMILES |
C1=CC=C2C(=C1)C(=CN2)C(=O)CO
|
|
| InChI |
InChI=1S/C10H9NO2/c12-6-10(13)8-5-11-9-4-2-1-3-7(8)9/h1-5,11-12H,6H2
|
|
| InChIKey |
IBLZDDPFMAFWKP-UHFFFAOYSA-N
|
|
| Synonyms |
2-Hydroxy-1-(1H-indol-3-yl)ethanone; 2400-51-3; 3-Glyceroindole; 2-Hydroxy-1-(3-indolyl)ethanone; Ethanone, 2-hydroxy-1-(1H-indol-3-yl)-; INDOLE-3-KETOL; Hydroxymethyl indol-3-yl ketone; 2-hydroxy-1-(1H-indol-3-yl)ethan-1-one; MFCD00466496; 2-hydroxy-1-(1H-indol-3-yl)-ethanone; BRN 0142060; 3-(Hydroxyacetyl)-1H-indole; 3-hydroxyacetylindole; 3-(Hydroxyacetyl)Indole; 3-(hydroxyacetyl)-indole; ChemDiv3_003333; 3-(hydroxylacetyl)-indole; Cambridge id 5106709; Oprea1_735053; MLS001049127; CHEMBL524903; SCHEMBL4056357; DTXSID80178731; CHEBI:173472; HMS1482H11; ZINC192311; 2-hydroxy-1-(indol-3-yl)ethanone; CAA40051; AC5698; STK386423; AKOS005431093; Hydroxymethyl indol-3-yl ketone, 8CI; IDI1_021243; NCGC00182562-01; BS-36792; SMR000386954; SY131003; CS-0450037; 2-Hydroxy-1-(1H-indol-3-yl)ethanone, 9CI
|
|
| CAS | 2400-51-3 | |
| PubChem CID | 200631 | |
| ChEMBL ID | CHEMBL524903 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 175.18 | ALogp: | 1.4 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 53.1 | Aromatic Rings: | 2 |
| Heavy Atoms: | 13 | QED Weighted: | 0.684 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.528 | MDCK Permeability: | 0.00000773 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.019 | 20% Bioavailability (F20%): | 0.006 |
| 30% Bioavailability (F30%): | 0.957 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.963 | Plasma Protein Binding (PPB): | 32.14% |
| Volume Distribution (VD): | 1.301 | Fu: | 62.16% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.895 | CYP1A2-substrate: | 0.192 |
| CYP2C19-inhibitor: | 0.208 | CYP2C19-substrate: | 0.107 |
| CYP2C9-inhibitor: | 0.053 | CYP2C9-substrate: | 0.632 |
| CYP2D6-inhibitor: | 0.198 | CYP2D6-substrate: | 0.625 |
| CYP3A4-inhibitor: | 0.096 | CYP3A4-substrate: | 0.187 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.416 | Half-life (T1/2): | 0.919 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.036 | Human Hepatotoxicity (H-HT): | 0.176 |
| Drug-inuced Liver Injury (DILI): | 0.873 | AMES Toxicity: | 0.356 |
| Rat Oral Acute Toxicity: | 0.948 | Maximum Recommended Daily Dose: | 0.335 |
| Skin Sensitization: | 0.762 | Carcinogencity: | 0.152 |
| Eye Corrosion: | 0.085 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.856 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005757 | 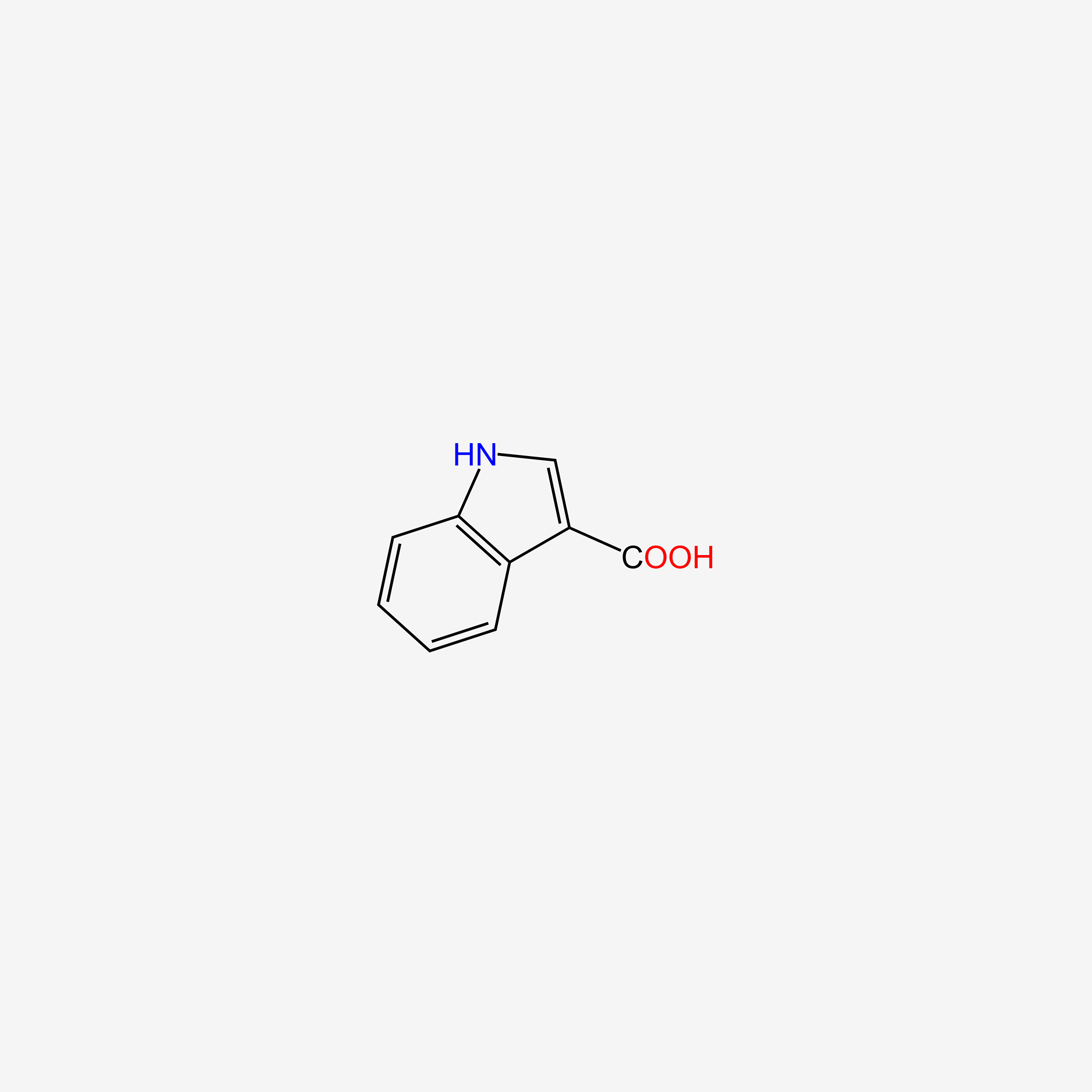 |
0.732 | D05EJG | 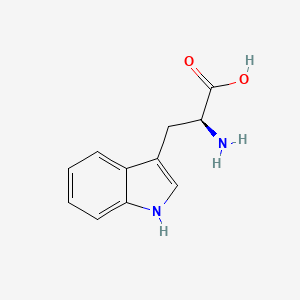 |
0.549 | ||
| ENC001345 | 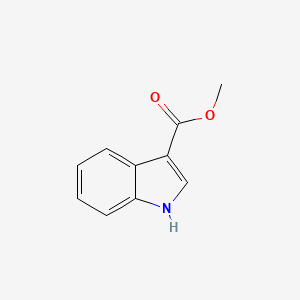 |
0.644 | D0K0KH | 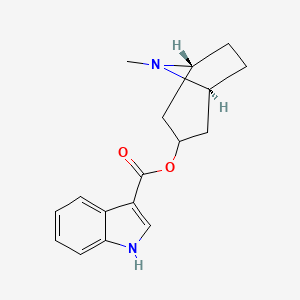 |
0.420 | ||
| ENC004871 | 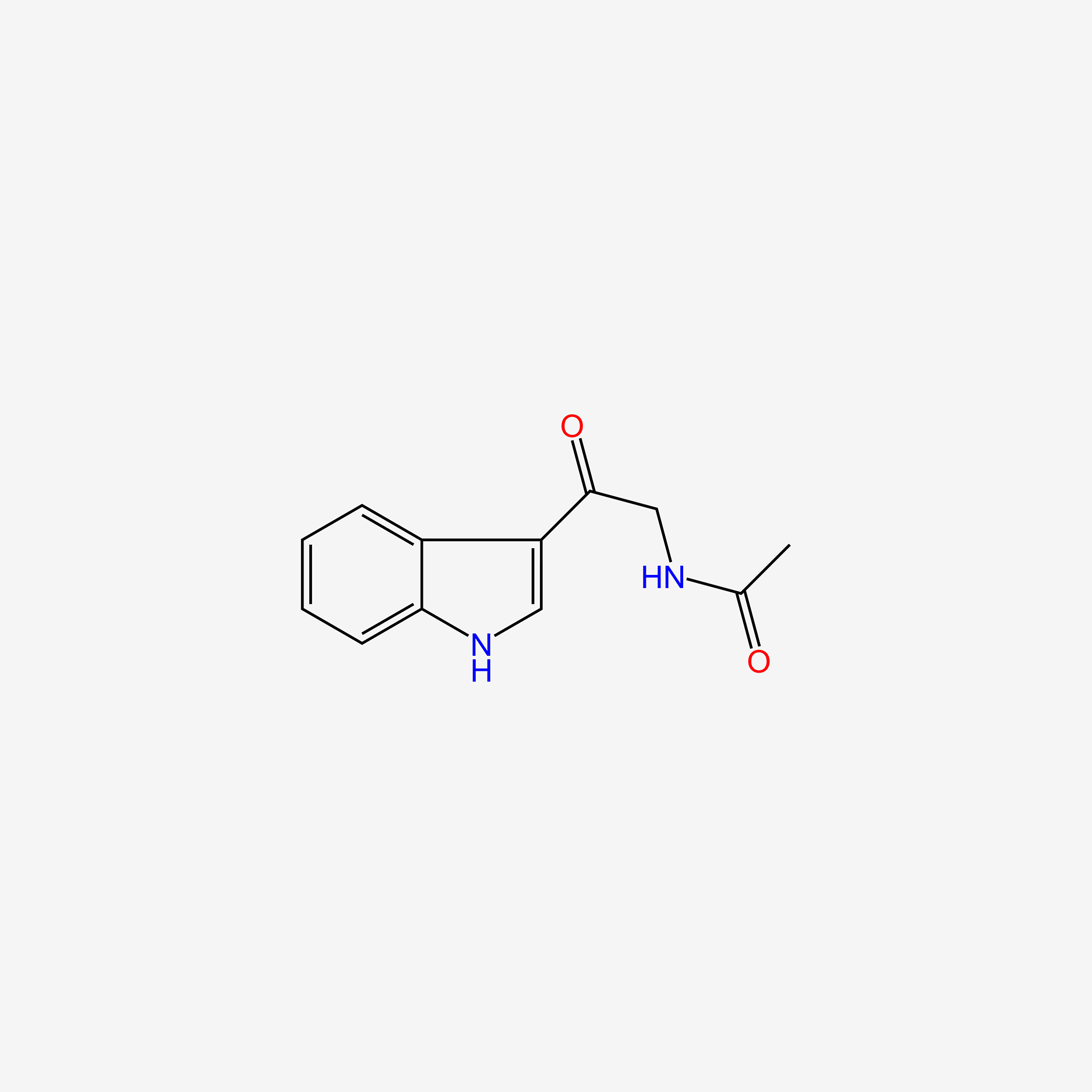 |
0.640 | D00YLW | 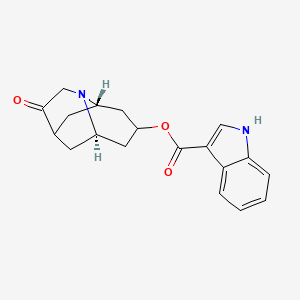 |
0.372 | ||
| ENC000043 | 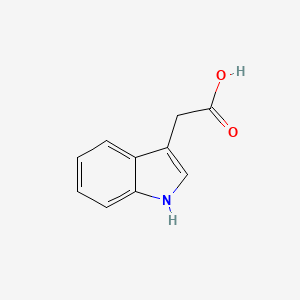 |
0.609 | D07HBX |  |
0.362 | ||
| ENC001448 | 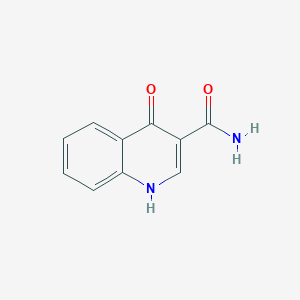 |
0.583 | D0W7WC | 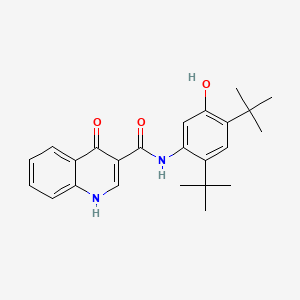 |
0.337 | ||
| ENC000363 | 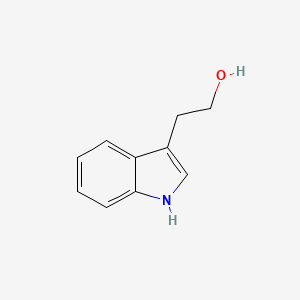 |
0.565 | D05OIS | 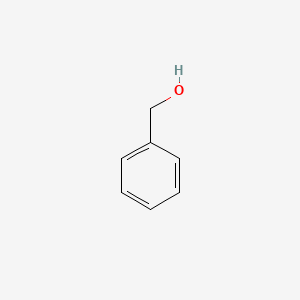 |
0.333 | ||
| ENC000140 | 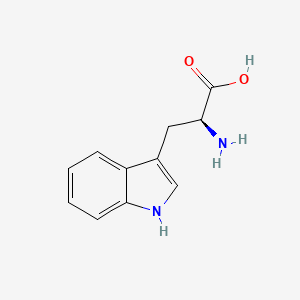 |
0.549 | D0R1CR | 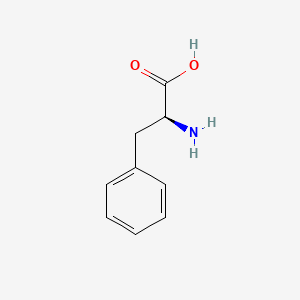 |
0.321 | ||
| ENC004706 | 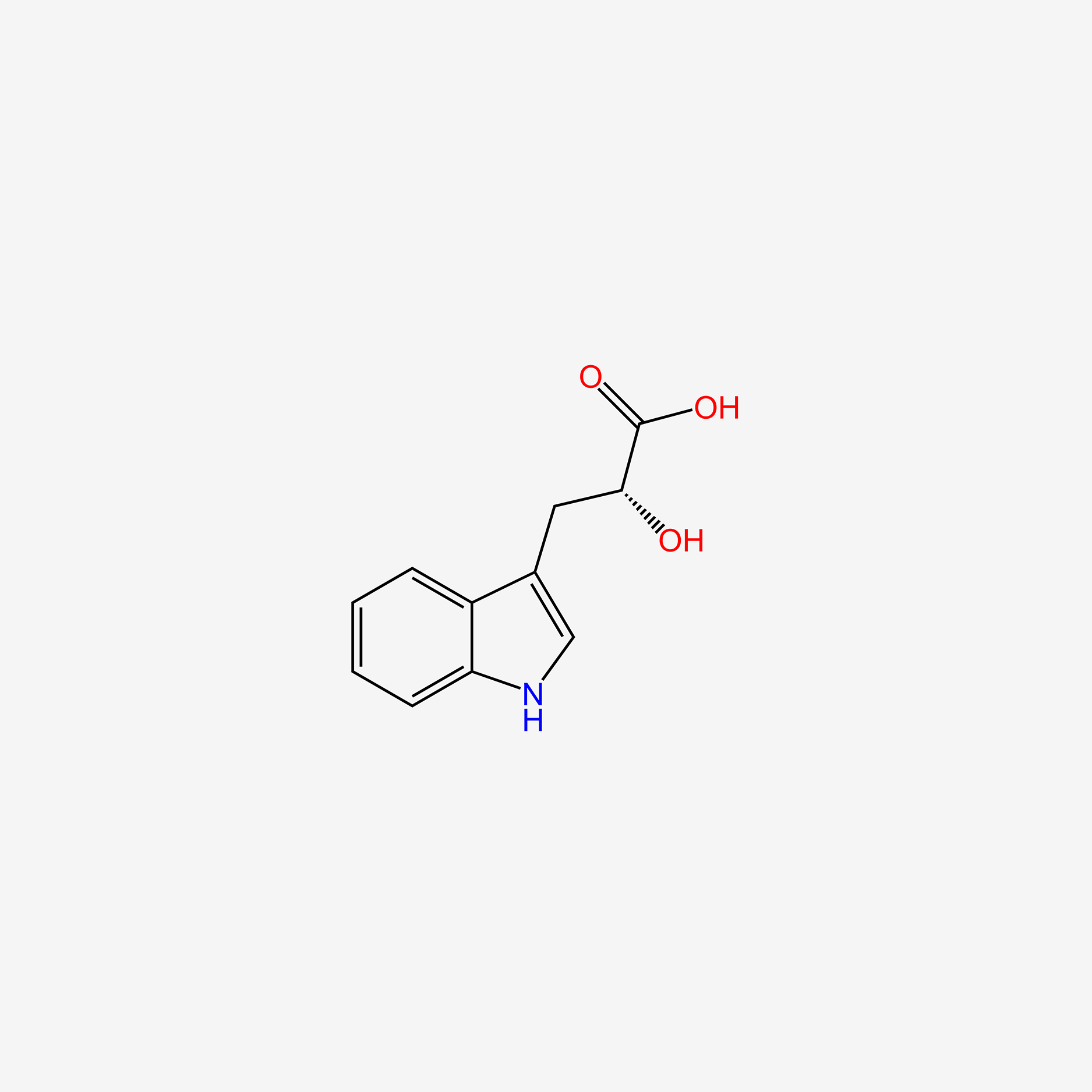 |
0.549 | D0X9RY | 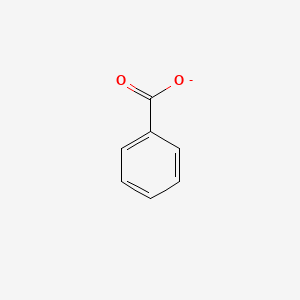 |
0.319 | ||
| ENC000341 |  |
0.533 | D0F5ZM | 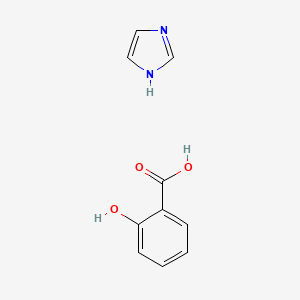 |
0.317 | ||
| ENC000042 | 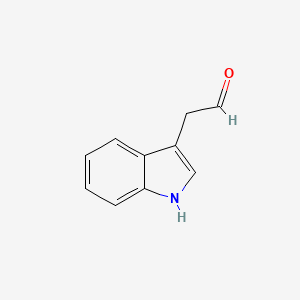 |
0.532 | D0GY5Z | 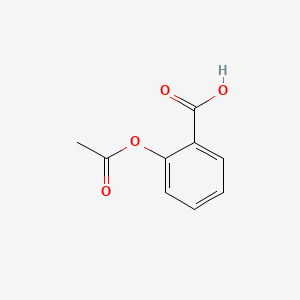 |
0.309 | ||