NPs Basic Information
|
Name |
3-Chloro-4-hydroxyphenylacetic acid
|
| Molecular Formula | C8H7ClO3 | |
| IUPAC Name* |
2-(3-chloro-4-hydroxyphenyl)acetic acid
|
|
| SMILES |
C1=CC(=C(C=C1CC(=O)O)Cl)O
|
|
| InChI |
InChI=1S/C8H7ClO3/c9-6-3-5(4-8(11)12)1-2-7(6)10/h1-3,10H,4H2,(H,11,12)
|
|
| InChIKey |
IYTUKSIOQKTZEG-UHFFFAOYSA-N
|
|
| Synonyms |
3-Chloro-4-hydroxyphenylacetic acid; 33697-81-3; 2-(3-chloro-4-hydroxyphenyl)acetic acid; (3-Chloro-4-hydroxyphenyl)acetic acid; 4-Hcpa; 3-Chloro-4-hydroxybenzeneacetic acid; Benzeneacetic acid, 3-chloro-4-hydroxy-; 4-Hydroxy-3-chlorophenylacetic acid; Acetic acid, (3-chloro-4-hydroxyphenyl)-; Kyselina 3-chlor-4-hydroxyfenyloctova; CHEMBL592010; CHEBI:47106; 3C4; m-Chloro-p-hydroxyphenylacetic acid; VUFB9649; EINECS 251-643-0; BRN 2096929; 3-CHLORO-4-HYDROXYPHENYLACETICACID; Kyselina 3-chlor-4-hydroxyfenyloctova [Czech]; 2h6b; 2-(3-chloro-4-hydroxy-phenyl)acetic acid; 3-10-00-00440 (Beilstein Handbook Reference); SCHEMBL377506; DTXSID60187381; ZINC407042; AMY28184; 3-chloro-4-hydroxylphenylacetic acid; BDBM50339588; MFCD00004349; 3-chloro-4-hydroxy-phenylacetic acid; 3-chloro-4-hydroxyphenyl-acetic acid; AKOS015890287; CS-W001146; PS-5822; {3-chloro-4-hydroxy-phenyl)acetic acid; AC-24895; 3-Chloro-4-hydroxyphenylacetic acid, 99%; DB-048474; A5988; FT-0639124; 3-CHLORO-4-HYDROXY PHENYL ACETIC ACID; EN300-78453; 2-(3-CHLORO-4-HYDROXYPHENYL)ACETICACID; C-4140; Q27120768; Z1205411134
|
|
| CAS | 33697-81-3 | |
| PubChem CID | 118534 | |
| ChEMBL ID | CHEMBL592010 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 186.59 | ALogp: | 1.5 |
| HBD: | 2 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 57.5 | Aromatic Rings: | 1 |
| Heavy Atoms: | 12 | QED Weighted: | 0.745 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.783 | MDCK Permeability: | 0.00003560 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.007 | 20% Bioavailability (F20%): | 0.009 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.119 | Plasma Protein Binding (PPB): | 89.43% |
| Volume Distribution (VD): | 0.236 | Fu: | 8.96% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.119 | CYP1A2-substrate: | 0.112 |
| CYP2C19-inhibitor: | 0.065 | CYP2C19-substrate: | 0.066 |
| CYP2C9-inhibitor: | 0.065 | CYP2C9-substrate: | 0.925 |
| CYP2D6-inhibitor: | 0.014 | CYP2D6-substrate: | 0.263 |
| CYP3A4-inhibitor: | 0.017 | CYP3A4-substrate: | 0.137 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.779 | Half-life (T1/2): | 0.92 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.202 |
| Drug-inuced Liver Injury (DILI): | 0.928 | AMES Toxicity: | 0.077 |
| Rat Oral Acute Toxicity: | 0.232 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.367 | Carcinogencity: | 0.382 |
| Eye Corrosion: | 0.257 | Eye Irritation: | 0.884 |
| Respiratory Toxicity: | 0.163 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
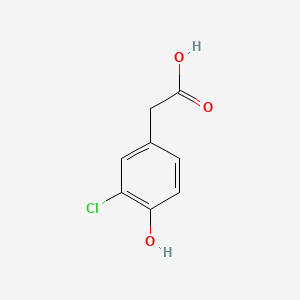
| ENC002317 | 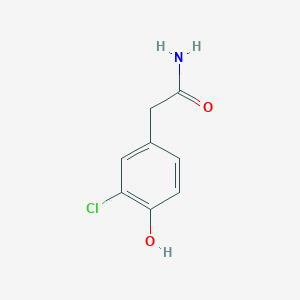 |
0.730 | D0C6OQ | 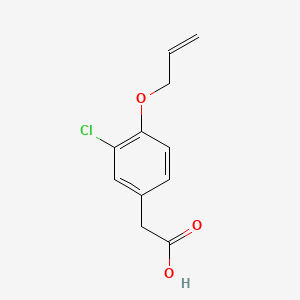 |
0.553 | ||
| ENC000035 |  |
0.684 | D08HVR | 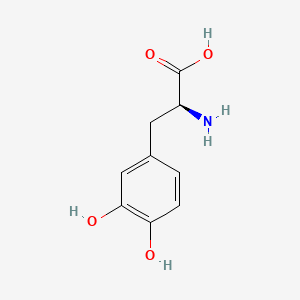 |
0.438 | ||
| ENC000006 | 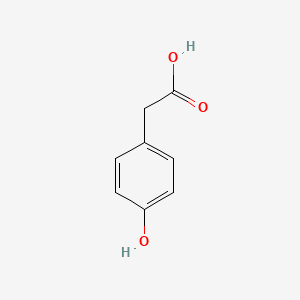 |
0.512 | D0BA6T | 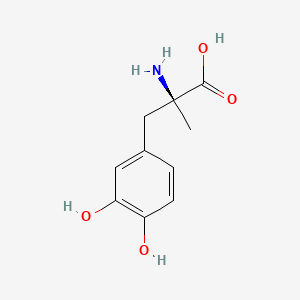 |
0.420 | ||
| ENC002095 | 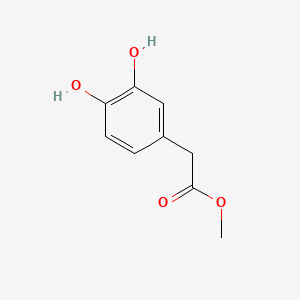 |
0.489 | D0C4YC | 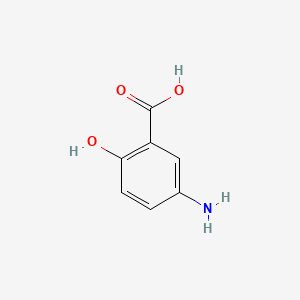 |
0.419 | ||
| ENC000409 | 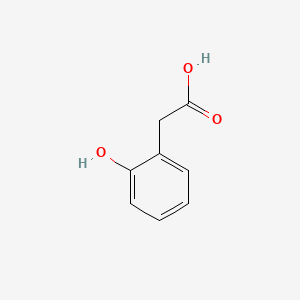 |
0.442 | D0P7JZ | 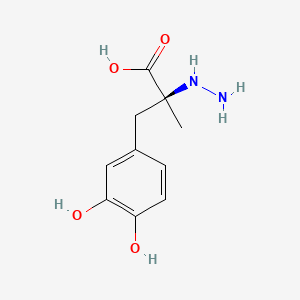 |
0.396 | ||
| ENC000127 | 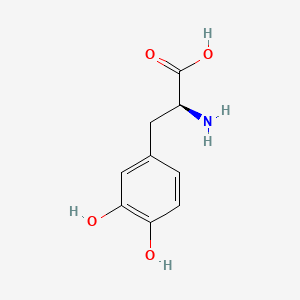 |
0.438 | D01WJL | 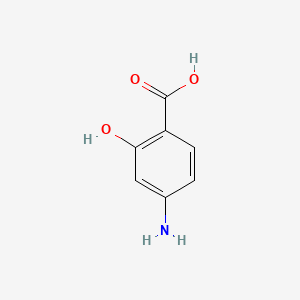 |
0.386 | ||
| ENC002136 | 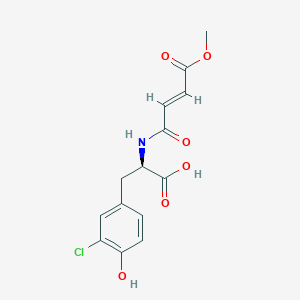 |
0.422 | D0U0OT | 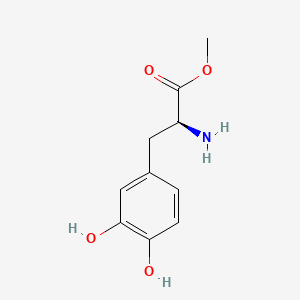 |
0.385 | ||
| ENC000103 | 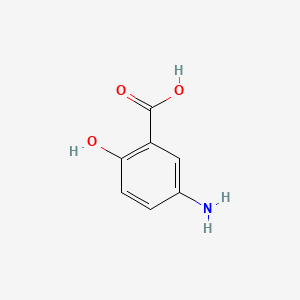 |
0.419 | D0T7OW | 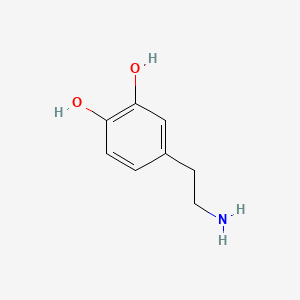 |
0.378 | ||
| ENC000002 | 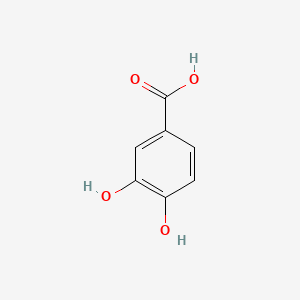 |
0.419 | D02AQY | 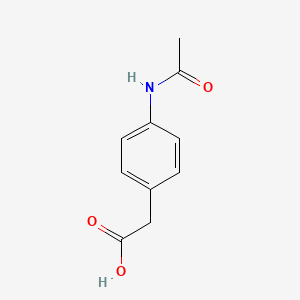 |
0.373 | ||
| ENC000097 | 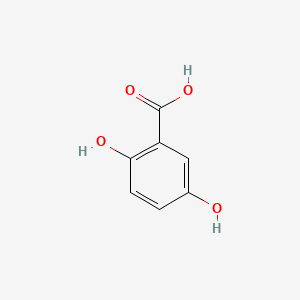 |
0.419 | D0V9EN | 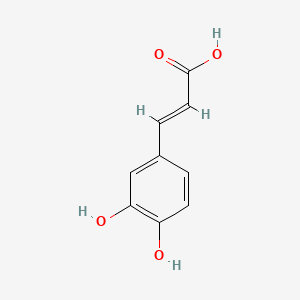 |
0.367 | ||