NPs Basic Information
|
Name |
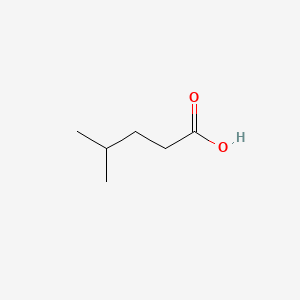
4-Methylpentanoic acid
|
| Molecular Formula | C6H12O2 | |
| IUPAC Name* |
4-methylpentanoic acid
|
|
| SMILES |
CC(C)CCC(=O)O
|
|
| InChI |
InChI=1S/C6H12O2/c1-5(2)3-4-6(7)8/h5H,3-4H2,1-2H3,(H,7,8)
|
|
| InChIKey |
FGKJLKRYENPLQH-UHFFFAOYSA-N
|
|
| Synonyms |
4-METHYLPENTANOIC ACID; 4-Methylvaleric acid; Isocaproic acid; 646-07-1; Isohexanoic acid; Pentanoic acid, 4-methyl-; Isobutylacetic acid; 4-methyl-pentanoic acid; Valeric acid, 4-methyl-; 4-METHYL VALERIC ACID; 4-Methyl-n-valeric acid; 4,4-Dimethylbutanoic acid; Isohexoic acid; FEMA No. 3463; 3-Methylbutane-1-carboxylic acid; NSC 4126; 4-methyl-Valeric acid; MFCD00002803; 4G4U8JA28T; CHEBI:74903; NSC-4126; Isohexanoate; Isohexoate; 4-Methylvalerate; 4-methyl-Valerate; 4-methyl-pentanoate; 4-methyl-n-valerate; 4,4-Dimethylbutanoate; 4MV; EINECS 211-464-0; BRN 1741912; UNII-4G4U8JA28T; AI3-04161; iso-caproic acid; 4-methyl pentanoic acid; 4,4-dimethylbutyric acid; bmse000625; 4-METHYLPENTANOICACID; SCHEMBL25603; 4-Methylvaleric acid, 99%; 4-02-00-00944 (Beilstein Handbook Reference); Butyl2,4-dichlorophenoxyacetate; WLN: QV2Y1&1; CHEMBL1230308; DTXSID8060951; NSC4126; HMS1732H05; ZINC391113; LMFA01020076; STL168053; 4-METHYLPENTANOIC ACID [FCC]; 4-METHYLPENTANOIC ACID [FHFI]; AKOS000121502; CS-W016598; DB03993; HY-W015882; 4-Methylvaleric Acid (Isocaproic Acid); LS-13180; SY001693; DB-003511; 4-Methylpentanoic acid, >=98%, FCC, FG; FT-0619146; M0457; M0750; S6278; EN300-16604; C21399; J-515800; Q19597905; Z56347204
|
|
| CAS | 646-07-1 | |
| PubChem CID | 12587 | |
| ChEMBL ID | CHEMBL1230308 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 116.16 | ALogp: | 1.4 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 8 | QED Weighted: | 0.612 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.638 | MDCK Permeability: | 0.00010853 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.002 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.836 | Plasma Protein Binding (PPB): | 63.06% |
| Volume Distribution (VD): | 0.247 | Fu: | 36.41% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.041 | CYP1A2-substrate: | 0.117 |
| CYP2C19-inhibitor: | 0.02 | CYP2C19-substrate: | 0.496 |
| CYP2C9-inhibitor: | 0.014 | CYP2C9-substrate: | 0.963 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.175 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.067 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.74 | Half-life (T1/2): | 0.806 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.113 |
| Drug-inuced Liver Injury (DILI): | 0.116 | AMES Toxicity: | 0.008 |
| Rat Oral Acute Toxicity: | 0.191 | Maximum Recommended Daily Dose: | 0.017 |
| Skin Sensitization: | 0.233 | Carcinogencity: | 0.144 |
| Eye Corrosion: | 0.98 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.06 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000351 | 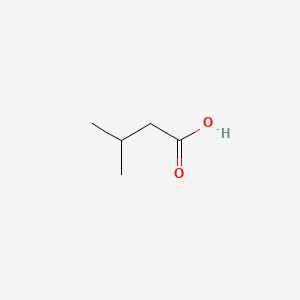 |
0.542 | D0R3QY |  |
0.433 | ||
| ENC000600 | 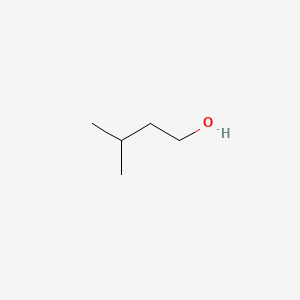 |
0.458 | D00WUF | 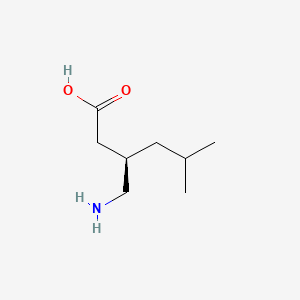 |
0.412 | ||
| ENC000018 | 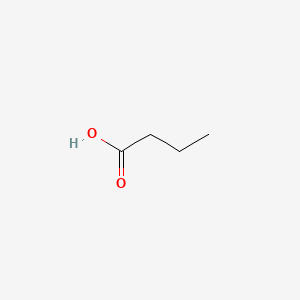 |
0.458 | D00ENY | 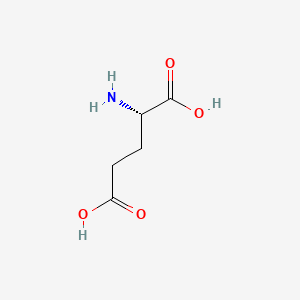 |
0.406 | ||
| ENC000149 | 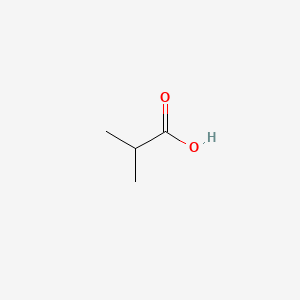 |
0.435 | D0EP8X | 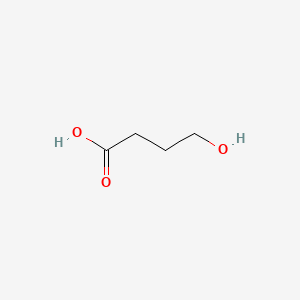 |
0.357 | ||
| ENC000603 | 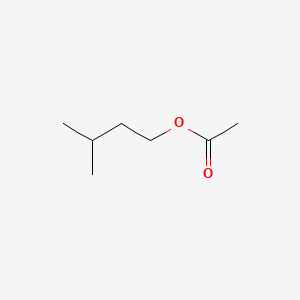 |
0.433 | D06VNK | 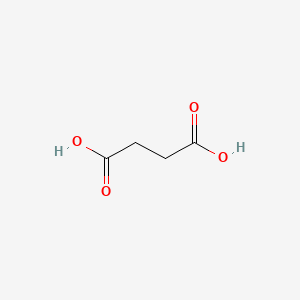 |
0.333 | ||
| ENC001015 | 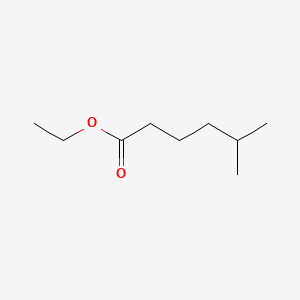 |
0.400 | D0Z0MG | 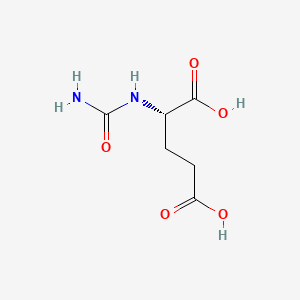 |
0.325 | ||
| ENC000231 | 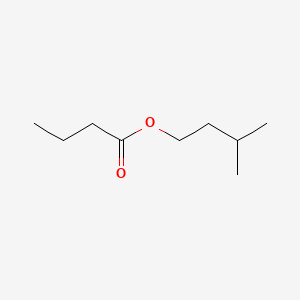 |
0.400 | D09PUL | 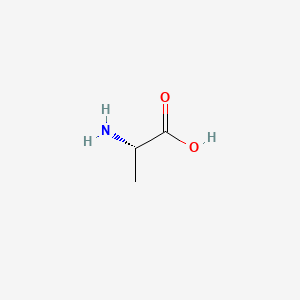 |
0.320 | ||
| ENC000677 | 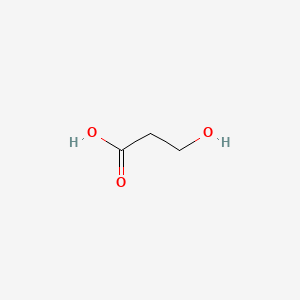 |
0.400 | D0Y3KG | 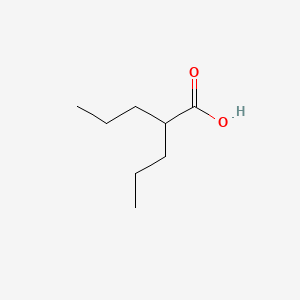 |
0.314 | ||
| ENC000227 | 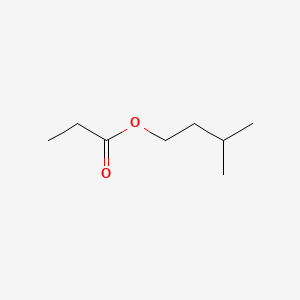 |
0.394 | D08QGD | 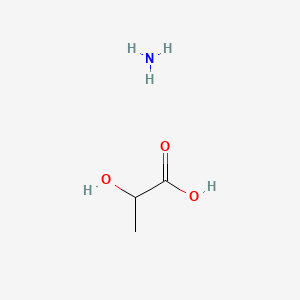 |
0.308 | ||
| ENC000685 | 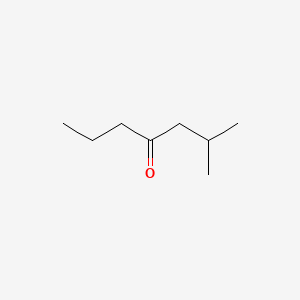 |
0.387 | D0Y7ZD | 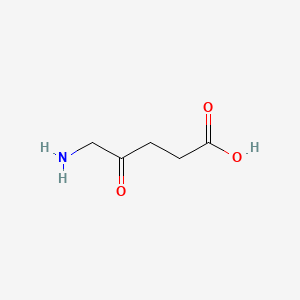 |
0.303 | ||