NPs Basic Information
|
Name |
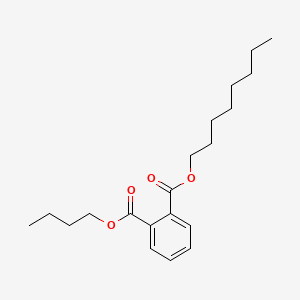
Butyl octyl phthalate
|
| Molecular Formula | C20H30O4 | |
| IUPAC Name* |
1-O-butyl 2-O-octyl benzene-1,2-dicarboxylate
|
|
| SMILES |
CCCCCCCCOC(=O)C1=CC=CC=C1C(=O)OCCCC
|
|
| InChI |
InChI=1S/C20H30O4/c1-3-5-7-8-9-12-16-24-20(22)18-14-11-10-13-17(18)19(21)23-15-6-4-2/h10-11,13-14H,3-9,12,15-16H2,1-2H3
|
|
| InChIKey |
MURWRBWZIMXKGC-UHFFFAOYSA-N
|
|
| Synonyms |
Butyl octyl phthalate; 84-78-6; Plasticizer OBP; Staflex BOP; Octyl butyl phthalate; Plasticizer BOP; Truflex OBP; 1,2-Benzenedicarboxylic acid, butyl octyl ester; PX 914; Phthalic acid, butyl octyl ester; 1-O-butyl 2-O-octyl benzene-1,2-dicarboxylate; NSC 69894; 1,2-Benzenedicarboxylic acid, 1-butyl 2-octyl ester; Phthalic acid, butyloctyl ester; Butyl octyl phthalate(Technical); Y52J9Q533Y; NSC-69894; EINECS 201-562-1; BRN 2289337; UNII-Y52J9Q533Y; 1, butyl octyl ester; SCHEMBL50451; Cbz-DL-3-Aminoisobutyricacid; 1-Butyl 2-octyl phthalate #; DTXSID2052578; AMY28401; NSC69894; MFCD00072261; Phthalic acid 1-butyl 2-octyl ester; ZINC95851714; AS-57441; 1-butyl 2-octyl benzene-1,2-dicarboxylate; CS-0134595; C13887; A928579; Q27294278
|
|
| CAS | 84-78-6 | |
| PubChem CID | 66540 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 334.4 | ALogp: | 6.9 |
| HBD: | 0 | HBA: | 4 |
| Rotatable Bonds: | 14 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 52.6 | Aromatic Rings: | 1 |
| Heavy Atoms: | 24 | QED Weighted: | 0.376 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.673 | MDCK Permeability: | 0.00002450 |
| Pgp-inhibitor: | 0.954 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.001 | 20% Bioavailability (F20%): | 0.999 |
| 30% Bioavailability (F30%): | 0.997 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.027 | Plasma Protein Binding (PPB): | 97.88% |
| Volume Distribution (VD): | 1.367 | Fu: | 1.61% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.735 | CYP1A2-substrate: | 0.205 |
| CYP2C19-inhibitor: | 0.782 | CYP2C19-substrate: | 0.058 |
| CYP2C9-inhibitor: | 0.407 | CYP2C9-substrate: | 0.711 |
| CYP2D6-inhibitor: | 0.564 | CYP2D6-substrate: | 0.073 |
| CYP3A4-inhibitor: | 0.527 | CYP3A4-substrate: | 0.079 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.377 | Half-life (T1/2): | 0.123 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.233 | Human Hepatotoxicity (H-HT): | 0.004 |
| Drug-inuced Liver Injury (DILI): | 0.289 | AMES Toxicity: | 0.005 |
| Rat Oral Acute Toxicity: | 0.002 | Maximum Recommended Daily Dose: | 0.005 |
| Skin Sensitization: | 0.912 | Carcinogencity: | 0.242 |
| Eye Corrosion: | 0.034 | Eye Irritation: | 0.987 |
| Respiratory Toxicity: | 0.041 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000164 | 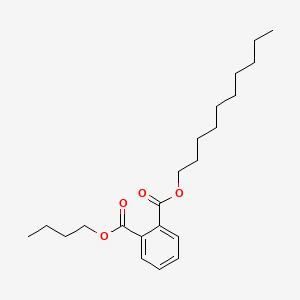 |
0.919 | D0K8CI | 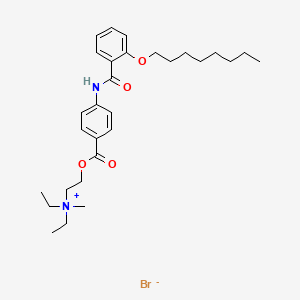 |
0.461 | ||
| ENC000291 | 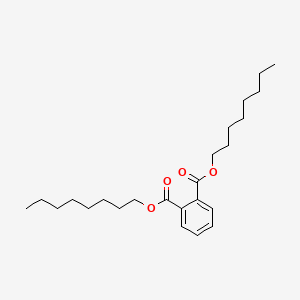 |
0.827 | D06ORU | 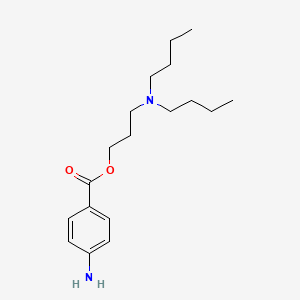 |
0.398 | ||
| ENC000090 | 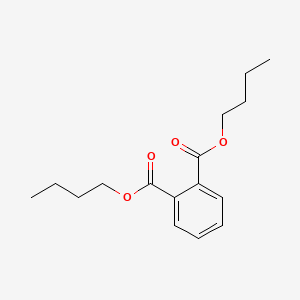 |
0.797 | D0OR6A | 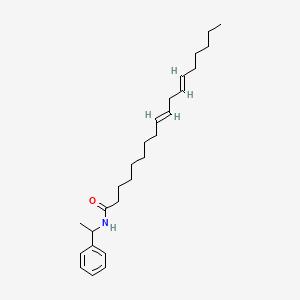 |
0.355 | ||
| ENC000156 | 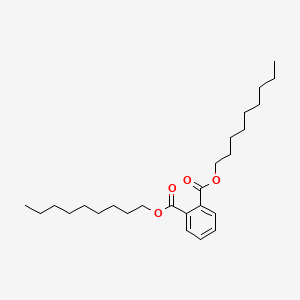 |
0.770 | D0G2KD | 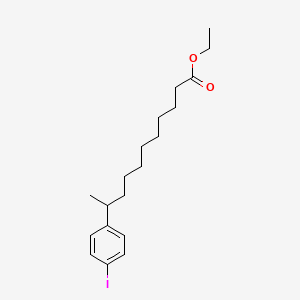 |
0.354 | ||
| ENC001801 | 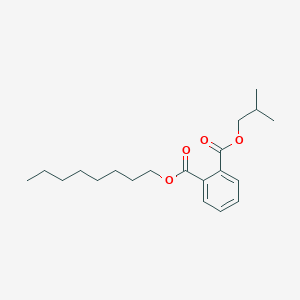 |
0.753 | D0AY9Q | 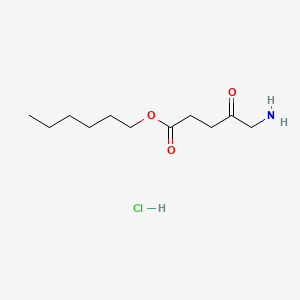 |
0.341 | ||
| ENC000158 | 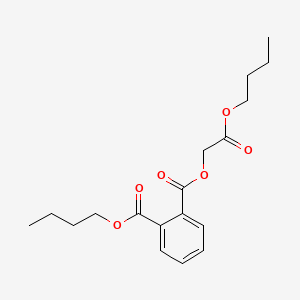 |
0.688 | D05ATI |  |
0.337 | ||
| ENC000616 | 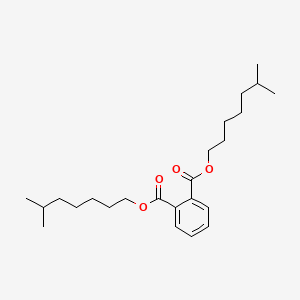 |
0.678 | D00MLW | 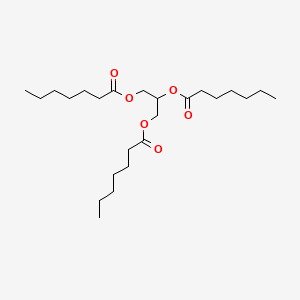 |
0.333 | ||
| ENC000157 | 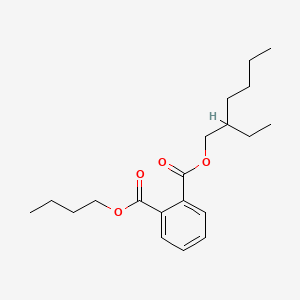 |
0.667 | D0H2SY | 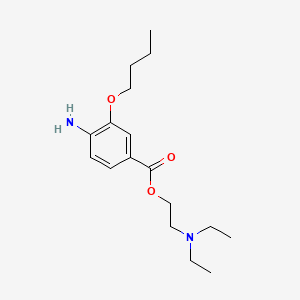 |
0.330 | ||
| ENC000300 | 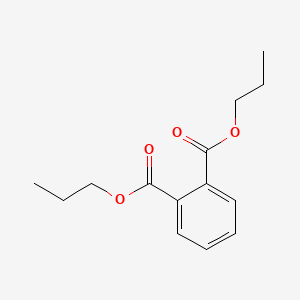 |
0.639 | D02MLW | 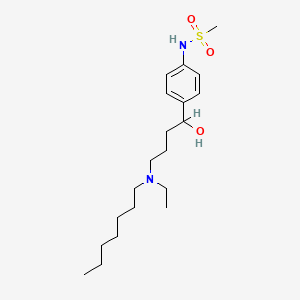 |
0.321 | ||
| ENC000293 | 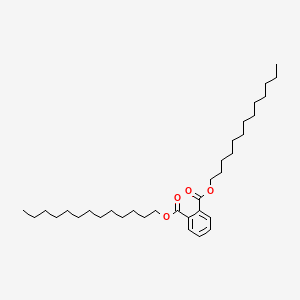 |
0.604 | D07UHS |  |
0.313 | ||