NPs Basic Information
|
Name |
Dinonyl phthalate
|
| Molecular Formula | C26H42O4 | |
| IUPAC Name* |
dinonyl benzene-1,2-dicarboxylate
|
|
| SMILES |
CCCCCCCCCOC(=O)C1=CC=CC=C1C(=O)OCCCCCCCCC
|
|
| InChI |
InChI=1S/C26H42O4/c1-3-5-7-9-11-13-17-21-29-25(27)23-19-15-16-20-24(23)26(28)30-22-18-14-12-10-8-6-4-2/h15-16,19-20H,3-14,17-18,21-22H2,1-2H3
|
|
| InChIKey |
DROMNWUQASBTFM-UHFFFAOYSA-N
|
|
| Synonyms |
DINONYL PHTHALATE; 84-76-4; Unimoll DN; Di-n-nonyl phthalate; Bisoflex 91; dinonyl benzene-1,2-dicarboxylate; Nonyl phthalate; 1,2-Benzenedicarboxylic acid, dinonyl ester; Bisoflex DNP; Phthalic acid, dinonyl ester; 1,2-dinonyl benzene-1,2-dicarboxylate; Dinonyl 1,2-benzenedicarboxylate; 90UCU78V8R; Phthalic acid, bis-nonyl ester; 68515-45-7; Bisoflex 91; Bisoflex DNP;Unimoll DN; Bisolflex 91; 1,2-Benzenedicarboxylic acid, 1,2-dinonyl ester; Ditrimethylhexyl phthalate; Phthalic Acid Dinonyl Ester; Di-n-nonylphthalate (DnNP); HSDB 365; EINECS 201-560-0; BRN 1916263; UNII-90UCU78V8R; Ceneg; Phthalic acid dinonyl; DSSTox_CID_8663; Diisononyl phthalate-[d4]; Dinonyl Phthalate, Technical; DSSTox_RID_82694; DSSTox_GSID_47966; 4-09-00-03183 (Beilstein Handbook Reference); SCHEMBL111972; CHEMBL3186308; DINONYL PHTHALATE [HSDB]; DTXSID9047966; Tox21_201040; MFCD00036237; STL280349; ZINC68589536; Dinonyl phthalate, mixture of isomers; AKOS015839921; CAS-84-76-4; NCGC00248904-01; NCGC00258593-01; 68648-92-0; BS-29460; FT-0625189; EN300-19868; Q27271347
|
|
| CAS | 84-76-4 | |
| PubChem CID | 6787 | |
| ChEMBL ID | CHEMBL3186308 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 418.6 | ALogp: | 10.1 |
| HBD: | 0 | HBA: | 4 |
| Rotatable Bonds: | 20 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 52.6 | Aromatic Rings: | 1 |
| Heavy Atoms: | 30 | QED Weighted: | 0.181 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.95 | MDCK Permeability: | 0.00001430 |
| Pgp-inhibitor: | 0.618 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.001 | 20% Bioavailability (F20%): | 1 |
| 30% Bioavailability (F30%): | 0.999 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.017 | Plasma Protein Binding (PPB): | 98.67% |
| Volume Distribution (VD): | 2.333 | Fu: | 1.18% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.135 | CYP1A2-substrate: | 0.172 |
| CYP2C19-inhibitor: | 0.46 | CYP2C19-substrate: | 0.048 |
| CYP2C9-inhibitor: | 0.107 | CYP2C9-substrate: | 0.86 |
| CYP2D6-inhibitor: | 0.393 | CYP2D6-substrate: | 0.039 |
| CYP3A4-inhibitor: | 0.355 | CYP3A4-substrate: | 0.043 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.217 | Half-life (T1/2): | 0.039 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.258 | Human Hepatotoxicity (H-HT): | 0.002 |
| Drug-inuced Liver Injury (DILI): | 0.219 | AMES Toxicity: | 0.004 |
| Rat Oral Acute Toxicity: | 0.002 | Maximum Recommended Daily Dose: | 0.005 |
| Skin Sensitization: | 0.955 | Carcinogencity: | 0.186 |
| Eye Corrosion: | 0.03 | Eye Irritation: | 0.986 |
| Respiratory Toxicity: | 0.061 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
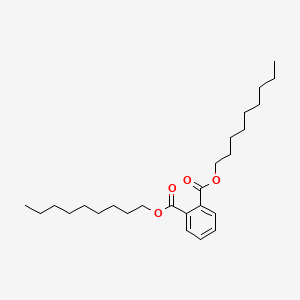
| ENC000291 | 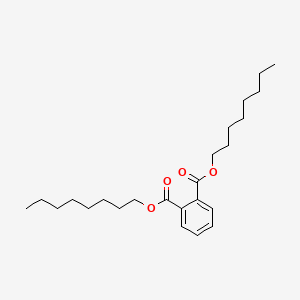 |
0.930 | D07ILQ |  |
0.431 | ||
| ENC000164 | 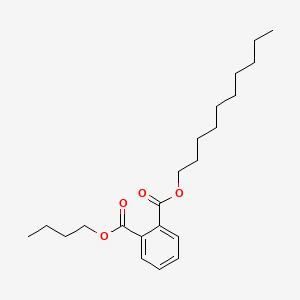 |
0.839 | D0Z5SM |  |
0.429 | ||
| ENC000293 |  |
0.782 | D00FGR | 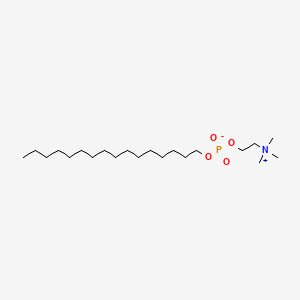 |
0.425 | ||
| ENC000669 | 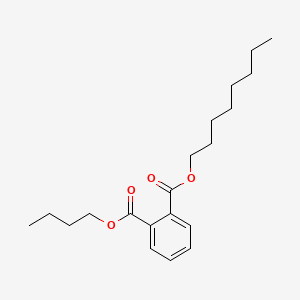 |
0.770 | D0K8CI | 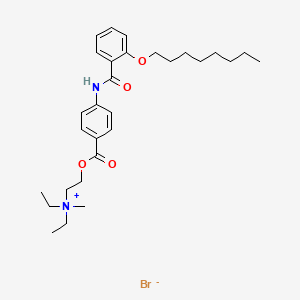 |
0.409 | ||
| ENC000090 | 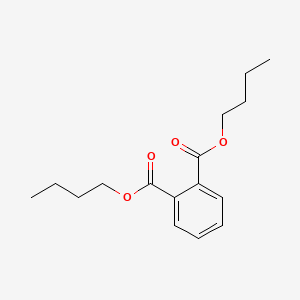 |
0.614 | D00MLW | 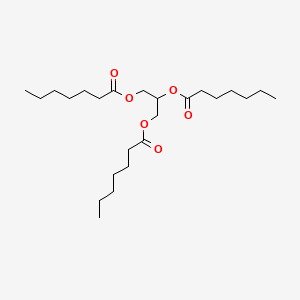 |
0.393 | ||
| ENC001801 | 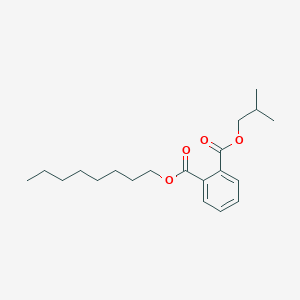 |
0.611 | D05ATI |  |
0.385 | ||
| ENC000601 | 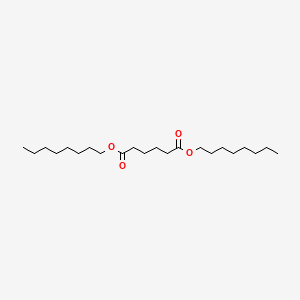 |
0.600 | D0P1RL | 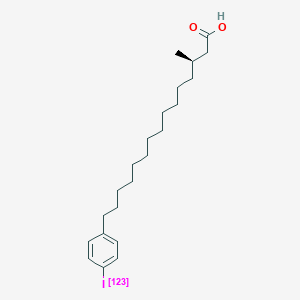 |
0.377 | ||
| ENC000616 | 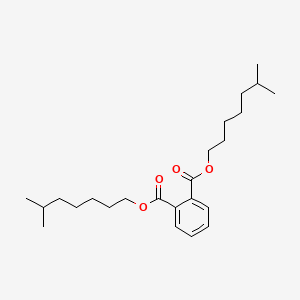 |
0.577 | D0T9TJ | 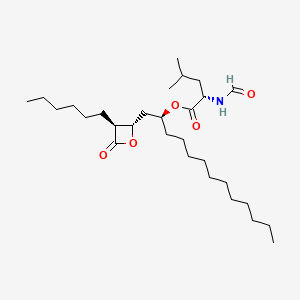 |
0.373 | ||
| ENC001234 | 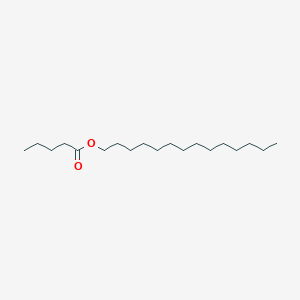 |
0.570 | D00AOJ | 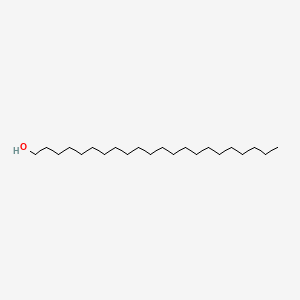 |
0.366 | ||
| ENC001218 |  |
0.551 | D0O1PH |  |
0.357 | ||