NPs Basic Information
|
Name |
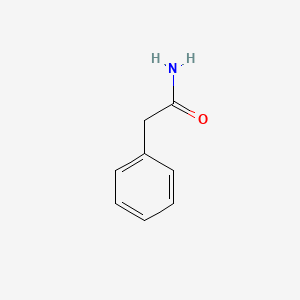
2-Phenylacetamide
|
| Molecular Formula | C8H9NO | |
| IUPAC Name* |
2-phenylacetamide
|
|
| SMILES |
C1=CC=C(C=C1)CC(=O)N
|
|
| InChI |
InChI=1S/C8H9NO/c9-8(10)6-7-4-2-1-3-5-7/h1-5H,6H2,(H2,9,10)
|
|
| InChIKey |
LSBDFXRDZJMBSC-UHFFFAOYSA-N
|
|
| Synonyms |
2-PHENYLACETAMIDE; 103-81-1; Benzeneacetamide; Phenylacetamide; alpha-Toluamide; Phenylacetic acid amide; alpha-Phenylacetamide; Acetamide, 2-phenyl-; Phenyl-beta-acetylamine; 2-phenyl-Acetamide; alpha-Toluimidic acid; .alpha.-Toluamide; .alpha.-Phenylacetamide; Phenyl-.beta.-acetylamine; CHEBI:16562; GNF-PF-1199; .alpha.-Toluimidic acid; 5R219M9TJF; CHEMBL347645; NSC-1877; MFCD00059193; NSC 1877; EINECS 203-147-0; phenacetamide; UNII-5R219M9TJF; phenyl acetamide; AI3-19421; benzene-acetamide; 2-phenyl acetamide; phenylacetimidic acid; Acetamide, 2-phenyl; (+)-benzeneacetamide; (-)-benzeneacetamide; beta-Phenyl-acetylamine; BENZENEDIACETAMIDE; (+/-)-benzeneacetamide; (alpha-)2-Phenylacetamide; 2-Phenylacetamide(alpha-); SCHEMBL25676; Phenylacetamide;alpha-Toluamide; DTXSID1059282; NSC1877; ZINC394644; ACT04844; BCP10408; BDBM50226209; s4440; .ALPHA.-PHENYLACETAMIDE [MI]; AKOS001215073; CS-W018983; HY-W018197; AC-24581; SY017094; DB-021492; A1882; FT-0613318; P0120; EN300-15619; C02505; Q-200316; Q27101974; Z33546508; F1084-0941; 46AFC744-1FF7-4816-B1C1-7C9995F369E8; N-O-NITROPHENYLSULFENYL-L-PROLINEDI(CYCLOHEXYL)AMMONIUMSALT
|
|
| CAS | 103-81-1 | |
| PubChem CID | 7680 | |
| ChEMBL ID | CHEMBL347645 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 135.16 | ALogp: | 0.5 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 43.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.648 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.732 | MDCK Permeability: | 0.00008920 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.96 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.076 |
| 30% Bioavailability (F30%): | 0.013 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.997 | Plasma Protein Binding (PPB): | 55.83% |
| Volume Distribution (VD): | 0.672 | Fu: | 47.21% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.286 | CYP1A2-substrate: | 0.111 |
| CYP2C19-inhibitor: | 0.059 | CYP2C19-substrate: | 0.107 |
| CYP2C9-inhibitor: | 0.031 | CYP2C9-substrate: | 0.191 |
| CYP2D6-inhibitor: | 0.009 | CYP2D6-substrate: | 0.231 |
| CYP3A4-inhibitor: | 0.017 | CYP3A4-substrate: | 0.236 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.825 | Half-life (T1/2): | 0.513 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.058 | Human Hepatotoxicity (H-HT): | 0.158 |
| Drug-inuced Liver Injury (DILI): | 0.777 | AMES Toxicity: | 0.468 |
| Rat Oral Acute Toxicity: | 0.015 | Maximum Recommended Daily Dose: | 0.008 |
| Skin Sensitization: | 0.409 | Carcinogencity: | 0.197 |
| Eye Corrosion: | 0.005 | Eye Irritation: | 0.726 |
| Respiratory Toxicity: | 0.015 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC005854 |  |
1.000 | D07ONP |  |
0.641 | ||
| ENC000054 |  |
0.697 | D0R1CR |  |
0.564 | ||
| ENC000218 | 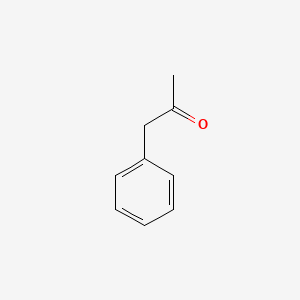 |
0.697 | D05OIS | 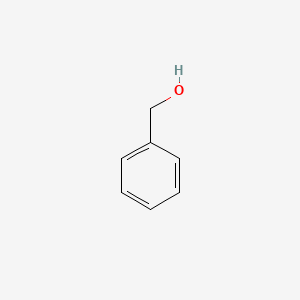 |
0.545 | ||
| ENC000208 | 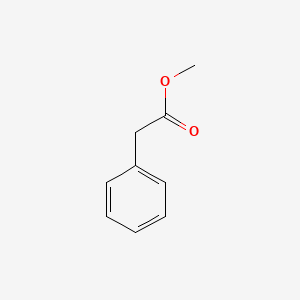 |
0.639 | D05BMG | 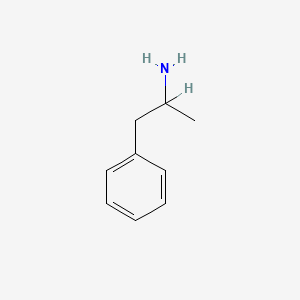 |
0.514 | ||
| ENC000130 | 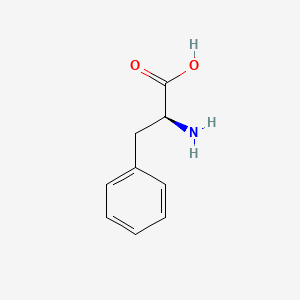 |
0.564 | D0T3LF | 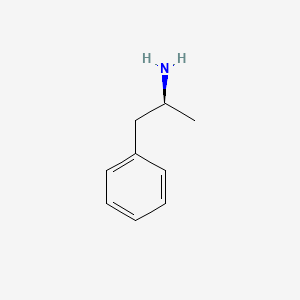 |
0.514 | ||
| ENC000076 | 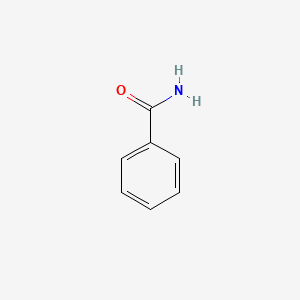 |
0.559 | D0P9AC | 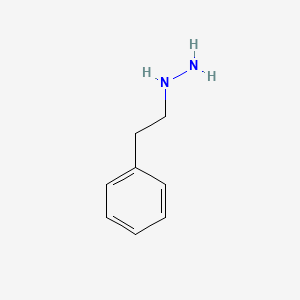 |
0.500 | ||
| ENC000004 | 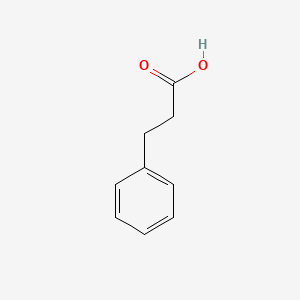 |
0.553 | D0P2GK | 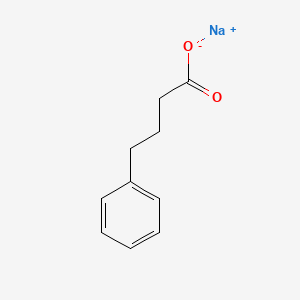 |
0.500 | ||
| ENC000308 | 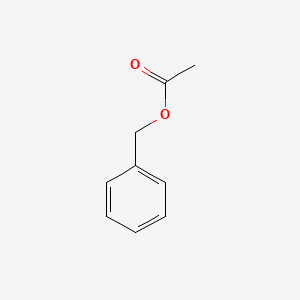 |
0.553 | D0U0RZ | 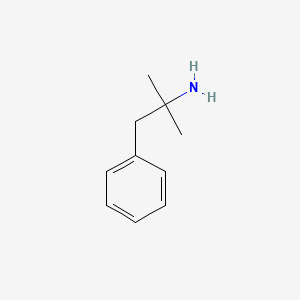 |
0.487 | ||
| ENC000203 |  |
0.545 | D00DZN |  |
0.477 | ||
| ENC000205 | 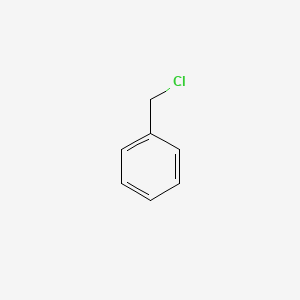 |
0.545 | D0X9RY | 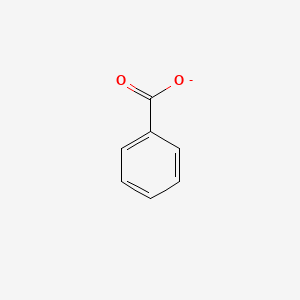 |
0.472 | ||