NPs Basic Information
|
Name |
Xanthurenic acid
|
| Molecular Formula | C10H7NO4 | |
| IUPAC Name* |
8-hydroxy-4-oxo-1H-quinoline-2-carboxylic acid
|
|
| SMILES |
C1=CC2=C(C(=C1)O)NC(=CC2=O)C(=O)O
|
|
| InChI |
InChI=1S/C10H7NO4/c12-7-3-1-2-5-8(13)4-6(10(14)15)11-9(5)7/h1-4,12H,(H,11,13)(H,14,15)
|
|
| InChIKey |
FBZONXHGGPHHIY-UHFFFAOYSA-N
|
|
| Synonyms |
xanthurenic acid; 59-00-7; 4,8-Dihydroxyquinoline-2-carboxylic acid; Xanthuric acid; 8-Hydroxykynurenic acid; 4,8-Dihydroxyquinaldic acid; Xanthurenate; 4,8-Dihydroxyquinaldinic acid; 8-hydroxy-4-oxo-1H-quinoline-2-carboxylic acid; 2-Quinolinecarboxylic acid, 4,8-dihydroxy-; NSC 401570; 4,8-Dihydroxy-2-quinolinecarboxylic acid; NSC401570; 58LAB1BG8J; QUINALDIC ACID, 4,8-DIHYDROXY-; CHEMBL312535; CHEBI:10072; NSC-401570; CCRIS 4429; 4,8-Dihydroxyquinoline-2-carboxylate; EINECS 200-410-1; UNII-58LAB1BG8J; BRN 0185954; Oxoxanthurenate; Xanthurate; 8-Hydroxykynurenate; 4-oxoxanthurenic acid; Spectrum_000253; 4,8-Dihydroxyquinaldate; Xanthurenic acid, 96%; Spectrum2_000158; Spectrum3_000143; Spectrum4_000117; Spectrum5_001562; 4,8-dihydroxy-Quinaldate; 4,8-Dihydroxyquinaldinate; Quinaldic acid,8-dihydroxy-; Oprea1_107134; BSPBio_001846; KBioGR_000474; KBioSS_000733; 4,8-dihydroxy-Quinaldic acid; BIDD:GT0640; DivK1c_000262; SCHEMBL379760; SPECTRUM1500754; XANTHURENIC ACID [MI]; SPBio_000296; 8-hydroxy-4-oxo-1,4-dihydroquinoline-2-carboxylic acid; HMS500N04; KBio1_000262; KBio2_000733; KBio2_003301; KBio2_005869; KBio3_001046; DTXSID90207728; NINDS_000262; HMS1921G08; HMS3885N22; WLN: T66 BNJ CVQ EQ JQ; ZINC8738372; 4,8-dihydroxyquinoline-2-carboxylic; BDBM50113313; CCG-38363; MFCD00006754; s4774; 4,8-Dihydroxy-2-quinolinecarboxylate; AKOS003237896; AKOS015894330; 4,8-dihydroxyquinolinium-2-carboxylate; CS-W015382; HY-W014666; SB72014; SDCCGMLS-0066616.P001; 2-Quinolinecarboxylic acid,8-dihydroxy-; IDI1_000262; NCGC00094846-01; NCGC00094846-02; NCGC00094846-03; NCGC00094846-04; NCGC00094846-05; AS-56782; BP-10912; DB-053303; AM20061355; FT-0631263; X0054; Quinoline-2-carboxylic acid, 4,8-dihydroxy-; C02470; X-1500; 4,8-Dihydroxy-quinoline-2-carboxylic acid anion; 059D007; A832107; SR-05000002445; CU-01000012491-3; Q5961262; SR-05000002445-1; BRD-K07327532-001-03-6; 4,8-Dihydroxy-quinoline-2-carboxylic acid(Xanthurenate); B5FF212D-76CB-4875-8A36-C67D8E69489C; Xanthurenate; 8-Hydroxykynurenic acid; 4,8-Dihydroxyquinaldic acid; 4KL
|
|
| CAS | 59-00-7 | |
| PubChem CID | 5699 | |
| ChEMBL ID | CHEMBL312535 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 205.17 | ALogp: | 0.9 |
| HBD: | 3 | HBA: | 5 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 86.6 | Aromatic Rings: | 2 |
| Heavy Atoms: | 15 | QED Weighted: | 0.655 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.239 | MDCK Permeability: | 0.00000463 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.08 | 20% Bioavailability (F20%): | 0.031 |
| 30% Bioavailability (F30%): | 0.978 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.208 | Plasma Protein Binding (PPB): | 47.95% |
| Volume Distribution (VD): | 0.483 | Fu: | 43.78% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.226 | CYP1A2-substrate: | 0.117 |
| CYP2C19-inhibitor: | 0.077 | CYP2C19-substrate: | 0.048 |
| CYP2C9-inhibitor: | 0.134 | CYP2C9-substrate: | 0.17 |
| CYP2D6-inhibitor: | 0.045 | CYP2D6-substrate: | 0.11 |
| CYP3A4-inhibitor: | 0.05 | CYP3A4-substrate: | 0.042 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.212 | Half-life (T1/2): | 0.896 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.011 | Human Hepatotoxicity (H-HT): | 0.26 |
| Drug-inuced Liver Injury (DILI): | 0.97 | AMES Toxicity: | 0.014 |
| Rat Oral Acute Toxicity: | 0.146 | Maximum Recommended Daily Dose: | 0.007 |
| Skin Sensitization: | 0.437 | Carcinogencity: | 0.028 |
| Eye Corrosion: | 0.007 | Eye Irritation: | 0.924 |
| Respiratory Toxicity: | 0.978 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
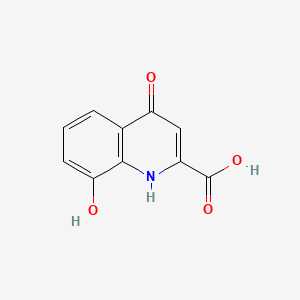
| ENC000997 | 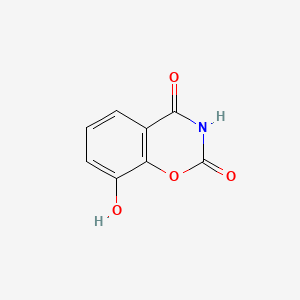 |
0.426 | D07HBX |  |
0.388 | ||
| ENC004685 | 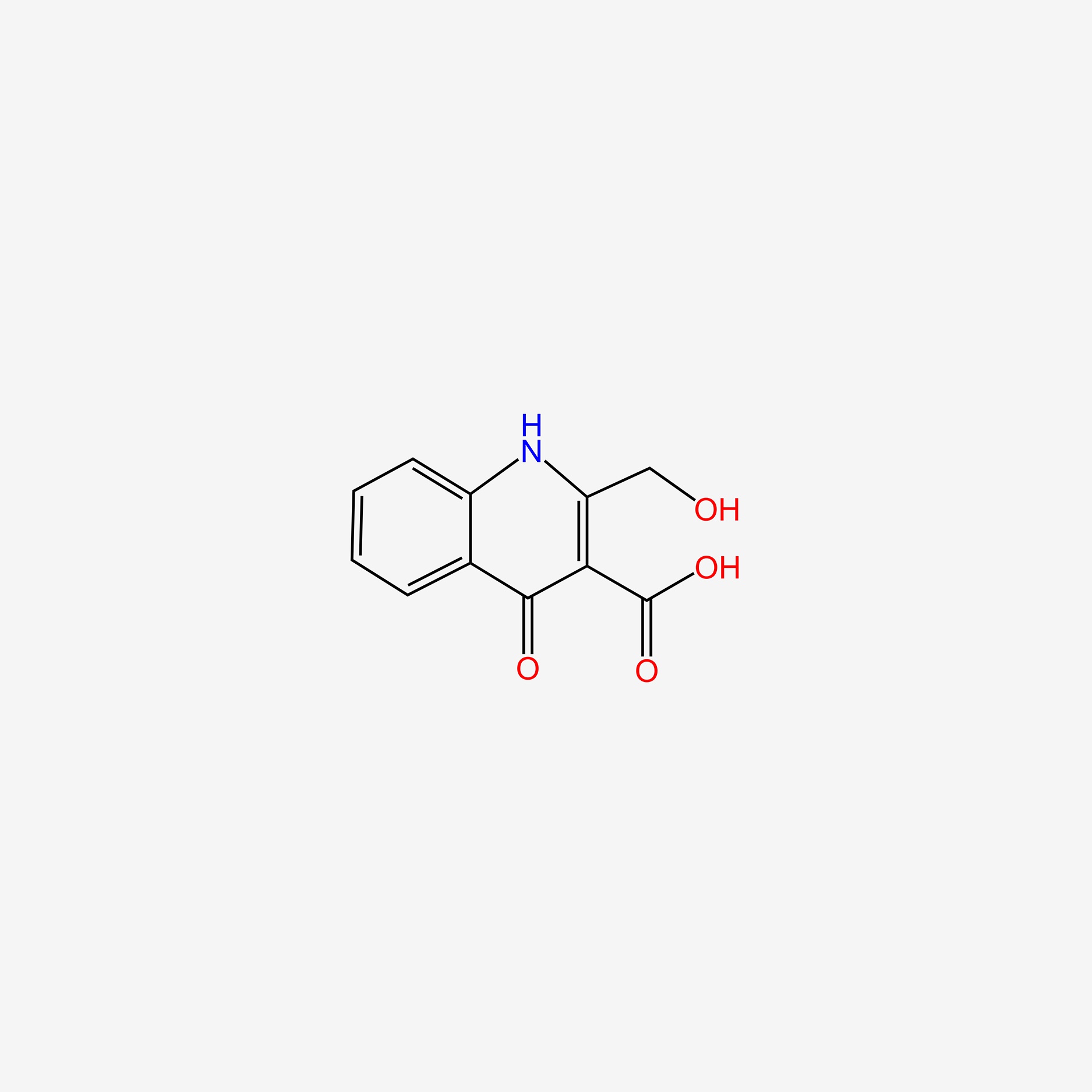 |
0.417 | D0C4YC |  |
0.346 | ||
| ENC000390 | 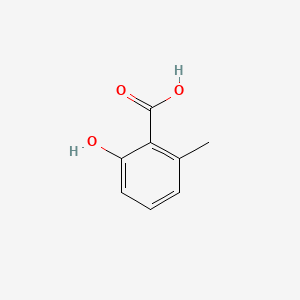 |
0.400 | D01WJL | 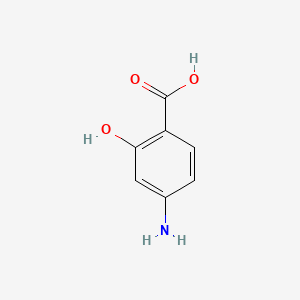 |
0.346 | ||
| ENC001447 | 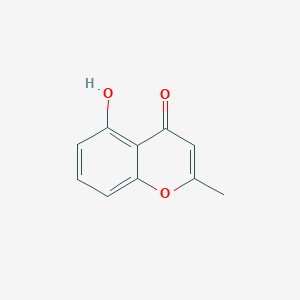 |
0.375 | D0F5ZM | 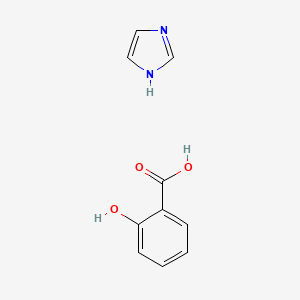 |
0.317 | ||
| ENC003945 | 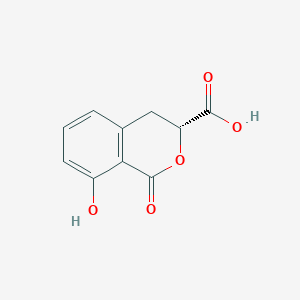 |
0.367 | D0Z1WA | 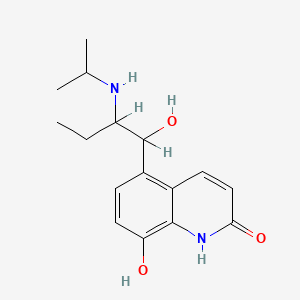 |
0.307 | ||
| ENC004829 | 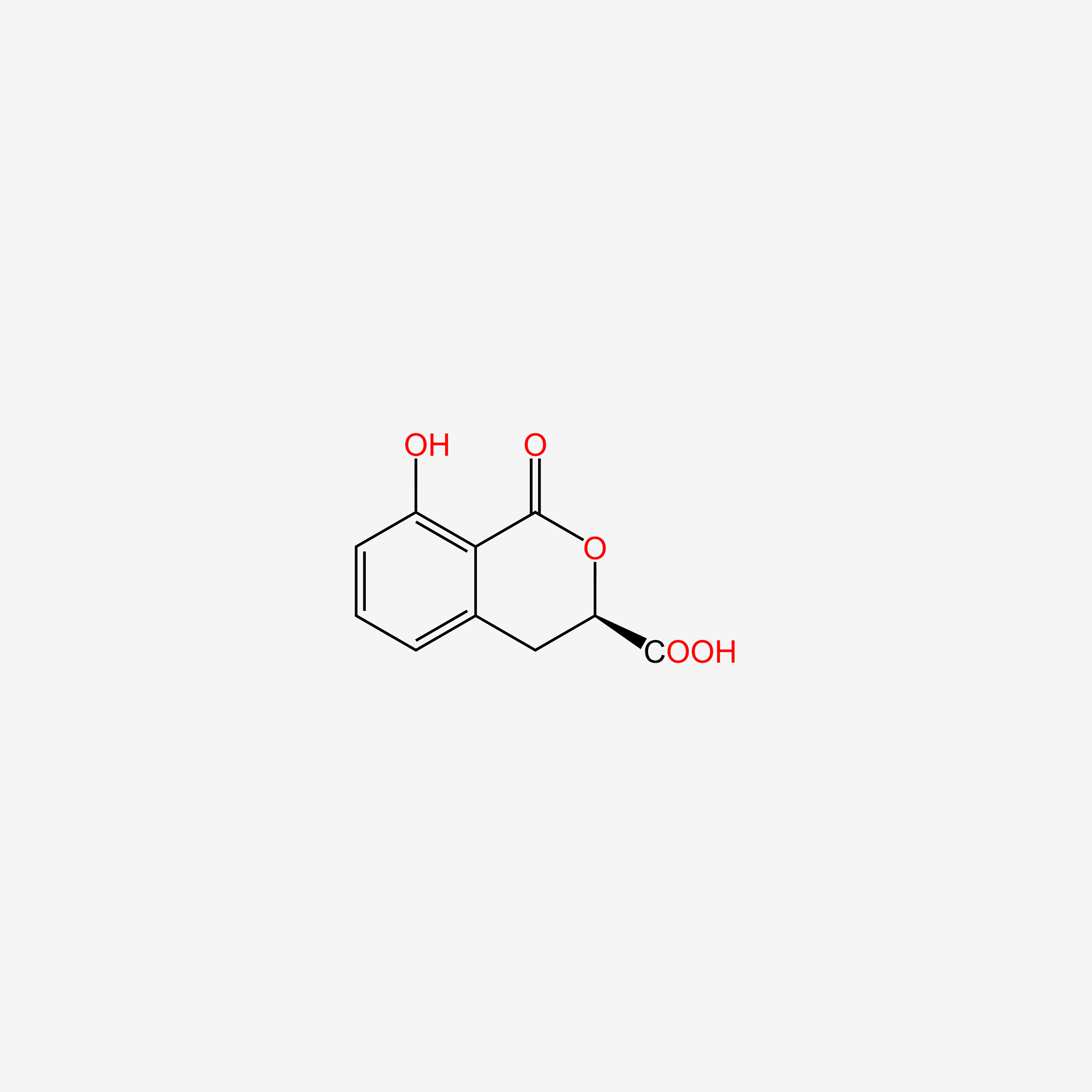 |
0.367 | D0Y0JH | 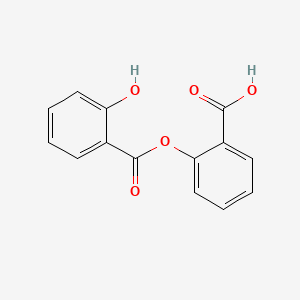 |
0.306 | ||
| ENC006051 |  |
0.366 | D08LFZ |  |
0.304 | ||
| ENC003390 |  |
0.361 | D0V9EN |  |
0.288 | ||
| ENC005347 | 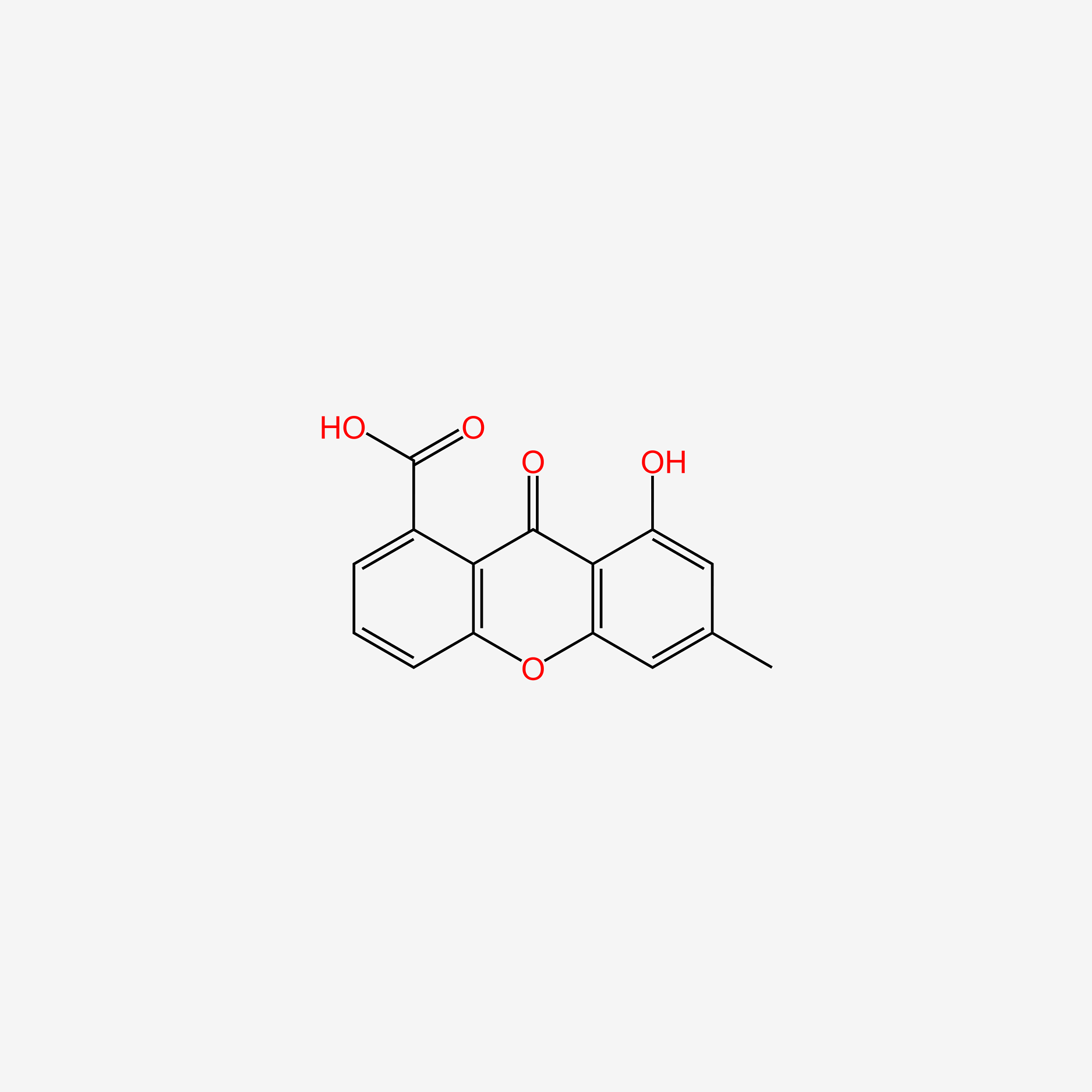 |
0.352 | D0N1WU | 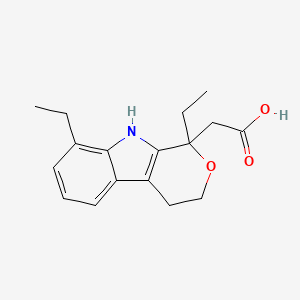 |
0.282 | ||
| ENC002362 | 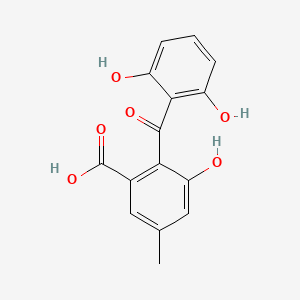 |
0.347 | D07JGT |  |
0.279 | ||