NPs Basic Information
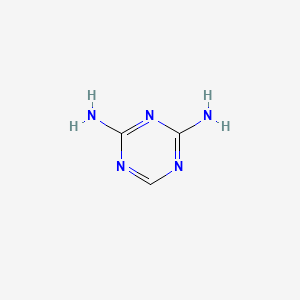
|
Name |
1,3,5-Triazine-2,4-diamine
|
| Molecular Formula | C3H5N5 | |
| IUPAC Name* |
1,3,5-triazine-2,4-diamine
|
|
| SMILES |
C1=NC(=NC(=N1)N)N
|
|
| InChI |
InChI=1S/C3H5N5/c4-2-6-1-7-3(5)8-2/h1H,(H4,4,5,6,7,8)
|
|
| InChIKey |
VZXTWGWHSMCWGA-UHFFFAOYSA-N
|
|
| Synonyms |
1,3,5-Triazine-2,4-diamine; 504-08-5; 2,4-Diamino-1,3,5-triazine; Formoguanamine; Guanamine; 4,6-Diamino-s-triazine; Diamino-s-triazine; 2,6-Diamino-s-triazine; 2,4-DIAMINO-S-TRIAZINE; s-Triazine, 2,4-diamino-; NSC 251; A6GN3JB7H4; NSC-251; EINECS 207-983-7; s-Triazine,4-diamino-; AI3-51263; UNII-A6GN3JB7H4; SCHEMBL65511; YSWG323; 1,5-Triazine-2,4-diamine; Fmoc-(3-aminophenyl)aceticacid; NSC251; DTXSID7074292; CHEBI:38071; [1,3,5]Triazine-2,4-diamine; ALBB-014718; ZINC1445475; MFCD00014598; STK256616; AKOS005174532; SB73700; AS-48301; CS-0110380; D2227; FT-0609961; D-2227; EN300-216072; F30229; A871573; Q27117366; ZZS
|
|
| CAS | 504-08-5 | |
| PubChem CID | 10435 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 111.11 | ALogp: | -0.7 |
| HBD: | 2 | HBA: | 5 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 90.7 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.472 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.008 | MDCK Permeability: | 0.00070076 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.963 |
| Human Intestinal Absorption (HIA): | 0.019 | 20% Bioavailability (F20%): | 0.028 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.688 | Plasma Protein Binding (PPB): | 9.68% |
| Volume Distribution (VD): | 1.005 | Fu: | 74.47% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.005 | CYP1A2-substrate: | 0.219 |
| CYP2C19-inhibitor: | 0.051 | CYP2C19-substrate: | 0.031 |
| CYP2C9-inhibitor: | 0.007 | CYP2C9-substrate: | 0.003 |
| CYP2D6-inhibitor: | 0 | CYP2D6-substrate: | 0.04 |
| CYP3A4-inhibitor: | 0.002 | CYP3A4-substrate: | 0.109 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.66 | Half-life (T1/2): | 0.929 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.049 | Human Hepatotoxicity (H-HT): | 0.999 |
| Drug-inuced Liver Injury (DILI): | 0.963 | AMES Toxicity: | 0.702 |
| Rat Oral Acute Toxicity: | 0.555 | Maximum Recommended Daily Dose: | 0.024 |
| Skin Sensitization: | 0.931 | Carcinogencity: | 0.708 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.957 |
| Respiratory Toxicity: | 0.921 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000011 | 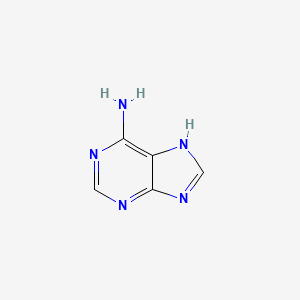 |
0.308 | D08IBS | 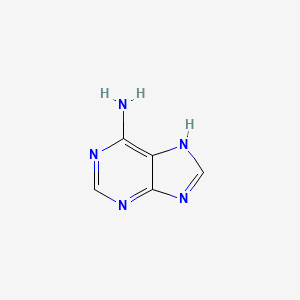 |
0.308 | ||
| ENC000467 | 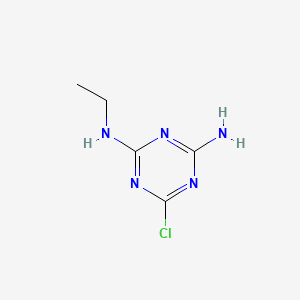 |
0.209 | D03FLC | 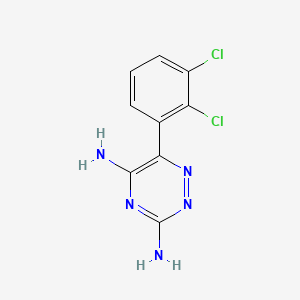 |
0.222 | ||
| ENC000635 | 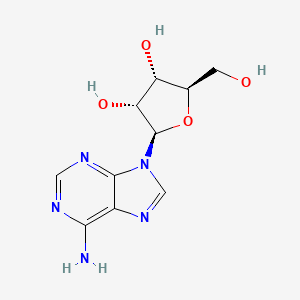 |
0.190 | D02ZXM | 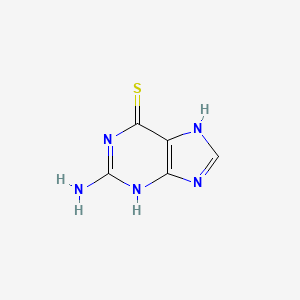 |
0.205 | ||
| ENC000599 | 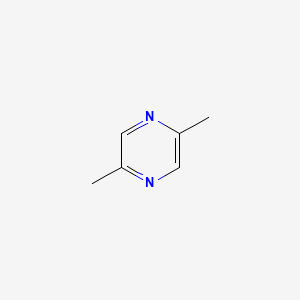 |
0.189 | D06QKV |  |
0.203 | ||
| ENC002667 | 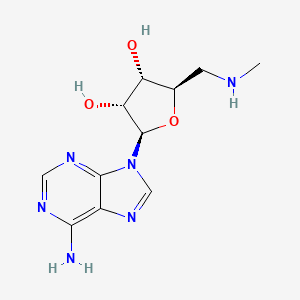 |
0.182 | D0X5XU | 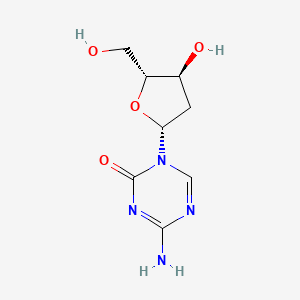 |
0.200 | ||
| ENC000577 | 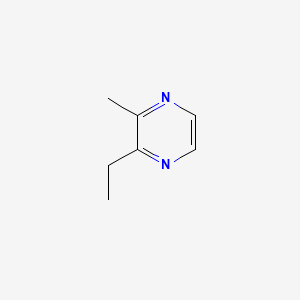 |
0.175 | D06HCP | 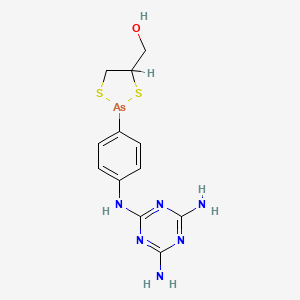 |
0.197 | ||
| ENC001225 |  |
0.167 | D0AO5H |  |
0.194 | ||
| ENC000728 |  |
0.163 | D0D9HW |  |
0.194 | ||
| ENC000721 | 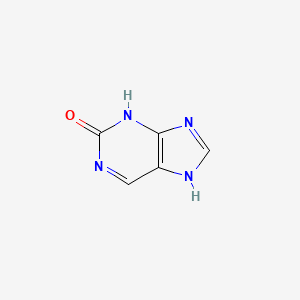 |
0.159 | D09FAZ | 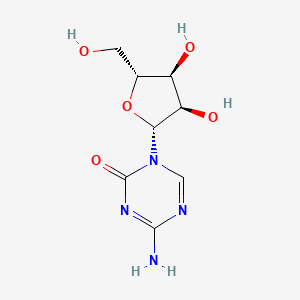 |
0.193 | ||
| ENC001351 | 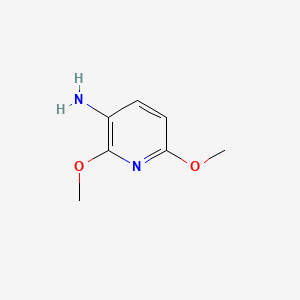 |
0.156 | D0NI0C | 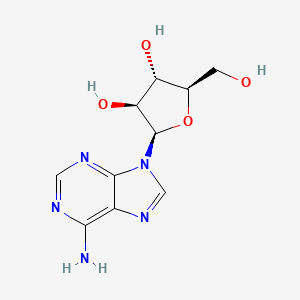 |
0.190 | ||