NPs Basic Information
|
Name |
2,5-Dimethylpyrazine
|
| Molecular Formula | C6H8N2 | |
| IUPAC Name* |
2,5-dimethylpyrazine
|
|
| SMILES |
CC1=CN=C(C=N1)C
|
|
| InChI |
InChI=1S/C6H8N2/c1-5-3-8-6(2)4-7-5/h3-4H,1-2H3
|
|
| InChIKey |
LCZUOKDVTBMCMX-UHFFFAOYSA-N
|
|
| Synonyms |
2,5-DIMETHYLPYRAZINE; 123-32-0; 2,5-Dimethyl pyrazine; Pyrazine, 2,5-dimethyl-; 2,5-Dimethyl-1,4-diazine; 2,5-Dimethylpiazine; 2,5-Dimethylparadiazine; NSC 49139; FEMA No. 3272; 2,5-Dimethyl-pyrazine; V99Y0MUY1Q; PYRAZINE,2,5-DIMETHYL; CHEBI:89762; MFCD00006147; NSC-49139; CCRIS 2929; 2,5-Dimethylpyrazine (natural); EINECS 204-618-3; UNII-V99Y0MUY1Q; Ketine; AI3-60303; 2.5-dimethylpyrazine; 2, 5-Dimethylpyrazine; pyrazine, 2,5-dimethyl; SCHEMBL82304; 2,5-Dimethylpyrazine, 98%; CHEMBL94709; ZINC3182; DTXSID6047652; FEMA 3272; WLN: T6N DNJ B1 E1; 2,5 and 2,6-dimethyl pyrazine; AMY23196; BCP08618; NSC49139; 2,5-DIMETHYLPYRAZINE [FCC]; 2,5-DIMETHYLPYRAZINE [FHFI]; 2,5-Dimethylpyrazine, >=98%, FG; AKOS003368403; CS-W019957; NCGC00184236-01; NCGC00184236-02; 2,5-Dimethylpyrazine, analytical standard; AC-10703; AS-17251; HY-34439; DB-003236; 2,5-Dimethylpyrazine (contains 2,6-isomer); D1526; D2171; FT-0610477; S3108; EN300-20206; 2,5-dimethylpyrazine and 2,6-dimethylpyrazine; 23D320; P19770; A805045; 2,5-Dimethyl Pyrazine; 2,5-Dimethyl-1,4-diazine; Q-100107; Q27161950; F0001-0364; 25R
|
|
| CAS | 123-32-0 | |
| PubChem CID | 31252 | |
| ChEMBL ID | CHEMBL94709 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 108.14 | ALogp: | 0.6 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 25.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 8 | QED Weighted: | 0.503 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.477 | MDCK Permeability: | 0.00003030 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.004 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.005 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.719 | Plasma Protein Binding (PPB): | 40.30% |
| Volume Distribution (VD): | 1.702 | Fu: | 68.15% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.301 | CYP1A2-substrate: | 0.747 |
| CYP2C19-inhibitor: | 0.026 | CYP2C19-substrate: | 0.72 |
| CYP2C9-inhibitor: | 0.003 | CYP2C9-substrate: | 0.441 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.822 |
| CYP3A4-inhibitor: | 0.008 | CYP3A4-substrate: | 0.265 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.66 | Half-life (T1/2): | 0.383 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.011 | Human Hepatotoxicity (H-HT): | 0.549 |
| Drug-inuced Liver Injury (DILI): | 0.306 | AMES Toxicity: | 0.025 |
| Rat Oral Acute Toxicity: | 0.166 | Maximum Recommended Daily Dose: | 0.036 |
| Skin Sensitization: | 0.832 | Carcinogencity: | 0.631 |
| Eye Corrosion: | 0.938 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.732 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
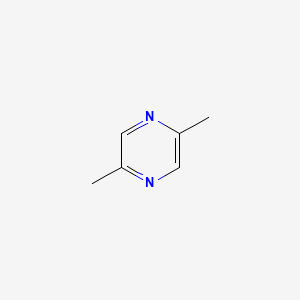
| ENC000240 |  |
0.333 | D02NJA | 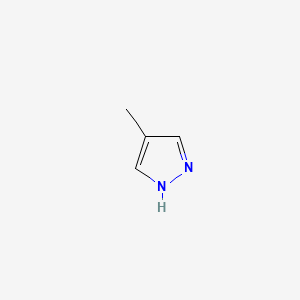 |
0.219 | ||
| ENC002316 | 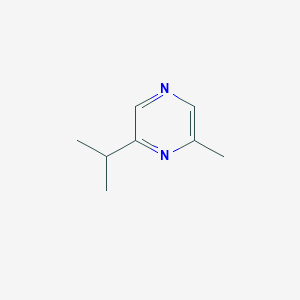 |
0.324 | D0S1NZ | 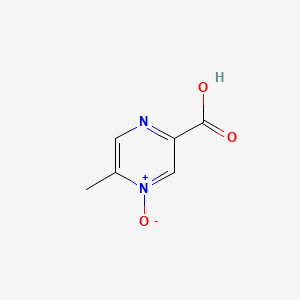 |
0.214 | ||
| ENC000577 | 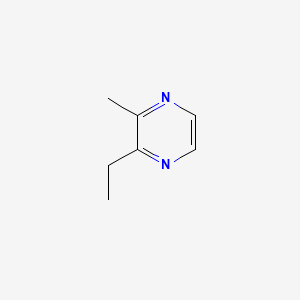 |
0.306 | D0U2CV |  |
0.208 | ||
| ENC000657 |  |
0.281 | D09MGR | 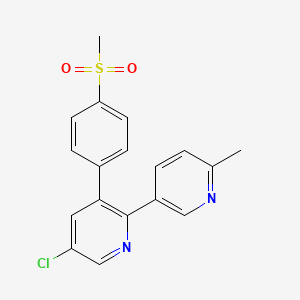 |
0.203 | ||
| ENC002253 | 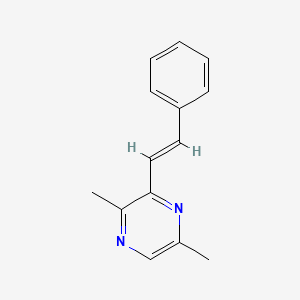 |
0.259 | D06PQT | 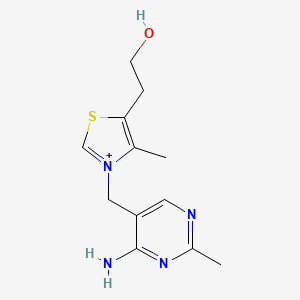 |
0.200 | ||
| ENC000650 | 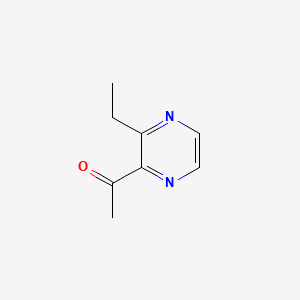 |
0.238 | D0V9YR | 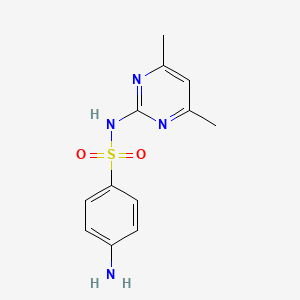 |
0.194 | ||
| ENC000092 | 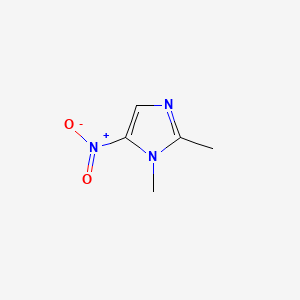 |
0.231 | D02LDV | 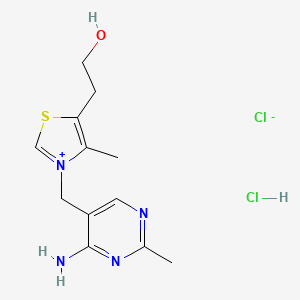 |
0.194 | ||
| ENC000239 | 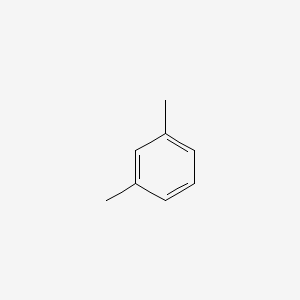 |
0.222 | D0V5IW | 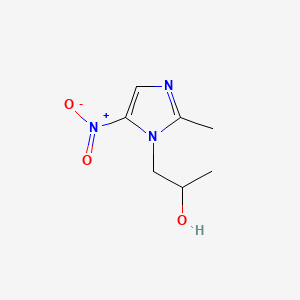 |
0.191 | ||
| ENC000233 | 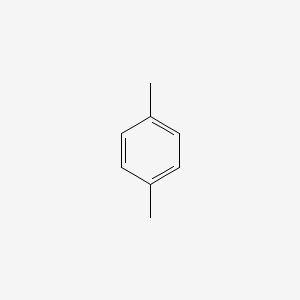 |
0.222 | D08IBS | 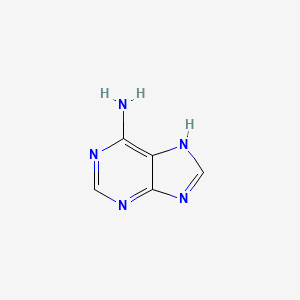 |
0.186 | ||
| ENC000179 |  |
0.222 | D04KYY |  |
0.186 | ||