NPs Basic Information
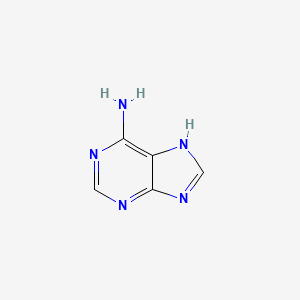
|
Name |
Adenine
|
| Molecular Formula | C5H5N5 | |
| IUPAC Name* |
7H-purin-6-amine
|
|
| SMILES |
C1=NC2=NC=NC(=C2N1)N
|
|
| InChI |
InChI=1S/C5H5N5/c6-4-3-5(9-1-7-3)10-2-8-4/h1-2H,(H3,6,7,8,9,10)
|
|
| InChIKey |
GFFGJBXGBJISGV-UHFFFAOYSA-N
|
|
| Synonyms |
adenine; 73-24-5; 1H-Purin-6-amine; 6-Aminopurine; 9H-Purin-6-amine; 7H-Purin-6-amine; Vitamin B4; Adenin; Adeninimine; Leuco-4; 6-Amino-1H-purine; 6-Amino-3H-purine; 6-Amino-7H-purine; 6-Amino-9H-purine; 1,6-Dihydro-6-iminopurine; 3,6-Dihydro-6-iminopurine; Purine, 6-amino-; USAF CB-18; 1H-Purine, 6-amino; Adenine [JAN]; ADE; 9H-Purine, 1,6-dihydro-6-imino-; 1H-Purine-6-amine; CCRIS 2556; AI3-50679; 3H-Purin-6(7H)-imine; NSC 14666; 9H-Purine-6-amine; 9H-purin-6-ylamine; 134434-48-3; CHEBI:16708; 1H-Purine, 6-amino-; MFCD00041790; NSC-14666; 6H-Purin-6-imine, 3,9-dihydro-, (Z)- (9CI); CHEMBL226345; 134434-49-4; 134454-76-5; 66224-66-6; JAC85A2161; 6H-Purin-6-imine, 1,7-dihydro-, (Z)- (9CI); 6H-Purin-6-imine, 1,9-dihydro-, (E)- (9CI); 6H-Purin-6-imine, 3,7-dihydro-, (Z)- (9CI); (Z)-3,9-Dihydro-6H-purin-6-imine; 134461-75-9; 71660-29-2; NCGC00094856-01; Pedatisectine B; DSSTox_CID_2557; DSSTox_RID_76627; DSSTox_GSID_22557; adenine-ring; CAS-73-24-5; Leucon (TN); Adenine (8CI); Adenine (JAN/USP); Adenine [USP:JAN]; SR-05000001754; 6-Aminopurine (Adenine); EINECS 200-796-1; 1H-purin-6(9H)-imine; 1H-Purin-6-amine (9CI); 3H-Purin-6-amine (9CI); UNII-JAC85A2161; 3h-adenine; 6-amino purine; 6-amino-Purine; purin-6-amine; 1jys; 1nli; 1wei; 2pqj; 3kpv; [3H]adenine; Adenine, 1; Adenine,(S); ALBB-005925; 7H-purin-6-ylamine; 71660-30-5; (S)-Norfluoxetine-d5; 9H-Purin-6-yl-amin; Adenine-[15N2]; Spectrum_001106; 2p8n; starbld0001134; 9H-Purin-6-yl-amine; ADENINE [VANDF]; Adenine-[8-13C]; SpecPlus_000535; ADENINE [INCI]; Adenine, >=99%; 9H-Purin-6-amine #; ADENINE [MI]; ADENINE [MART.]; Spectrum2_000583; Spectrum3_000616; Spectrum4_001891; Spectrum5_000542; ADENINE [USP-RS]; ADENINE [WHO-DD]; 6-Aminopurine;Vitamin B4; bmse000060; bmse000861; bmse000995; Epitope ID:140097; Adenine, cell culture grade; SCHEMBL8110; Oprea1_057274; US9138393, Adenine; US9144538, Adenine; BSPBio_002152; KBioGR_002447; KBioGR_002562; KBioSS_001586; KBioSS_002571; ZINC882; MLS001066342; DivK1c_006631; SPECTRUM1500807; SPBio_000426; ADENINE [EP MONOGRAPH]; ADENINE [USP IMPURITY]; GTPL4788; ADENINE [USP MONOGRAPH]; 9H-Purine,6-dihydro-6-imino-; DTXSID6022557; BDBM33218; KBio1_001575; KBio2_001586; KBio2_002562; KBio2_004154; KBio2_005130; KBio2_006722; KBio2_007698; KBio3_001652; KBio3_003040; 1,9-Dihydro-6H-purin-6-imine; Adenine 100 microg/mL in Water; cMAP_000085; 7H-Purin-6-amine, min. 95%; BCPP000433; BDBM181146; HMS1921I14; HMS2092K20; HMS2269I04; Pharmakon1600-01500807; BCP02865; HY-B0152; NSC14666; VCA70030; Tox21_111348; Tox21_302108; BBL007925; CCG-38506; NSC757793; s1981; STK387542; WLN: T56 BM DN FN HNJ IZ; AKOS000118903; AKOS005171607; Tox21_111348_1; AC-2028; AM83908; BCP9000233; CS-1984; DB00173; NSC-757793; SDCCGMLS-0066584.P001; NCGC00094856-02; NCGC00094856-03; NCGC00094856-05; NCGC00255120-01; 1217770-71-2; AS-11841; BL008313; NCI60_000998; SMR000471871; ADENOSINE IMPURITY A [EP IMPURITY]; SBI-0052324.P002; Adenine, Vetec(TM) reagent grade, >=99%; DB-013503; A0149; FT-0620943; FT-0656198; EN300-21472; 73A245; Adenine, suitable for cell culture, BioReagent; C00147; D00034; P50008; Q15277; Z-1043; AB00052833-18; AB00052833-19; AB00052833_20; AB00052833_22; AB00052833_23; AB00052833_24; A935233; Q-200595; SR-05000001754-1; SR-05000001754-2; W-106856; Adenine, BioReagent, plant cell culture tested, >=99%; Adenine, European Pharmacopoeia (EP) Reference Standard; F0001-1848; Z104498572; 6379C0E0-C1BB-4087-96C5-1DE281B8EA4C; Adenine, United States Pharmacopeia (USP) Reference Standard; Adenine, Pharmaceutical Secondary Standard; Certified Reference Material
|
|
| CAS | 73-24-5 | |
| PubChem CID | 190 | |
| ChEMBL ID | CHEMBL226345 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 135.13 | ALogp: | -0.1 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 80.5 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.546 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.072 | MDCK Permeability: | 0.00000429 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.993 |
| Human Intestinal Absorption (HIA): | 0.024 | 20% Bioavailability (F20%): | 0.828 |
| 30% Bioavailability (F30%): | 0.379 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.288 | Plasma Protein Binding (PPB): | 11.43% |
| Volume Distribution (VD): | 1.006 | Fu: | 79.68% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.13 | CYP1A2-substrate: | 0.536 |
| CYP2C19-inhibitor: | 0.16 | CYP2C19-substrate: | 0.058 |
| CYP2C9-inhibitor: | 0.015 | CYP2C9-substrate: | 0.347 |
| CYP2D6-inhibitor: | 0.395 | CYP2D6-substrate: | 0.077 |
| CYP3A4-inhibitor: | 0.029 | CYP3A4-substrate: | 0.224 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.729 | Half-life (T1/2): | 0.925 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.031 | Human Hepatotoxicity (H-HT): | 0.937 |
| Drug-inuced Liver Injury (DILI): | 0.973 | AMES Toxicity: | 0.346 |
| Rat Oral Acute Toxicity: | 0.983 | Maximum Recommended Daily Dose: | 0.061 |
| Skin Sensitization: | 0.928 | Carcinogencity: | 0.032 |
| Eye Corrosion: | 0.021 | Eye Irritation: | 0.976 |
| Respiratory Toxicity: | 0.979 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000352 | 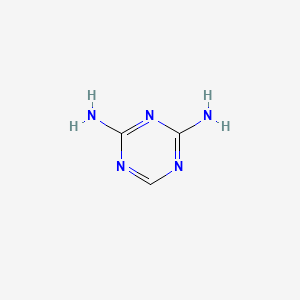 |
0.308 | D08IBS | 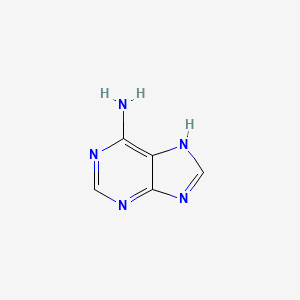 |
1.000 | ||
| ENC000721 | 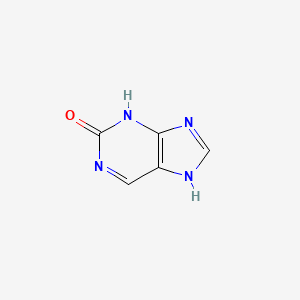 |
0.289 | D03OIW | 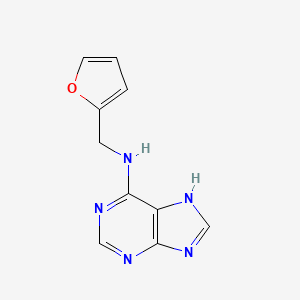 |
0.426 | ||
| ENC000635 | 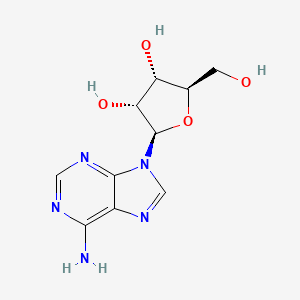 |
0.262 | D07QCE | 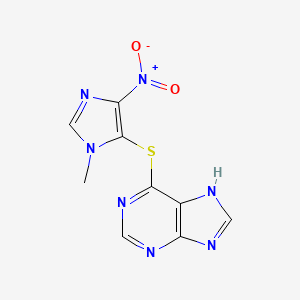 |
0.383 | ||
| ENC002667 | 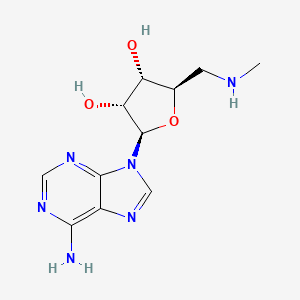 |
0.250 | D09UZO |  |
0.349 | ||
| ENC000322 | 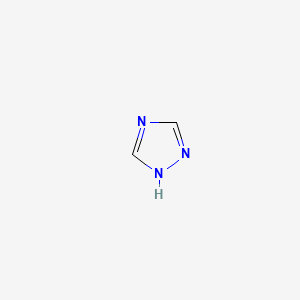 |
0.189 | D02ZXM | 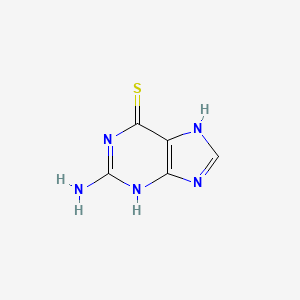 |
0.333 | ||
| ENC000599 | 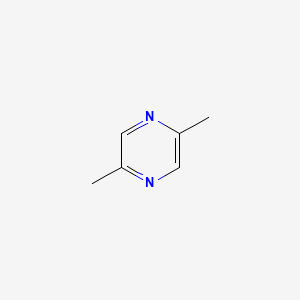 |
0.186 | D0D9HW | 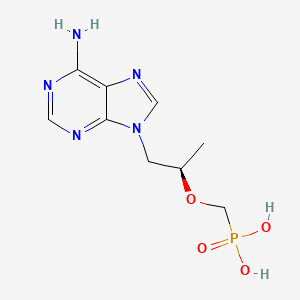 |
0.266 | ||
| ENC000784 | 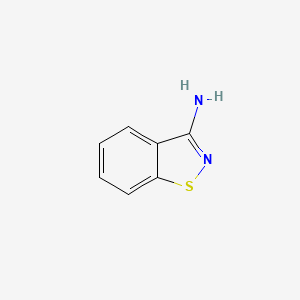 |
0.184 | D06IAR | 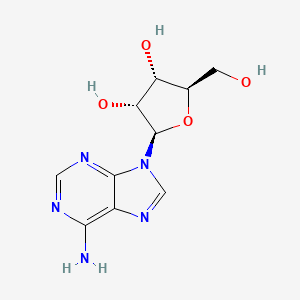 |
0.262 | ||
| ENC005790 | 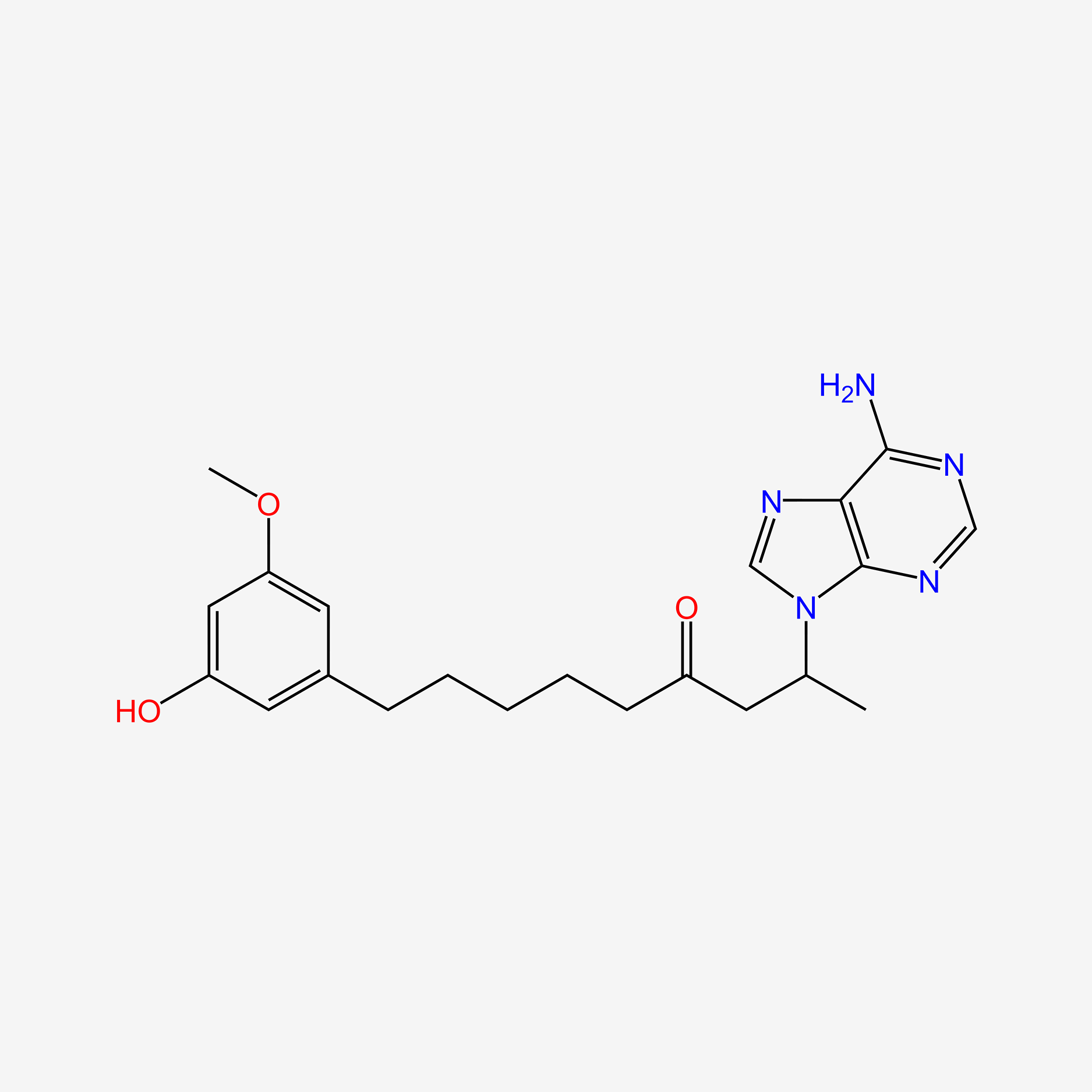 |
0.181 | D0NI0C | 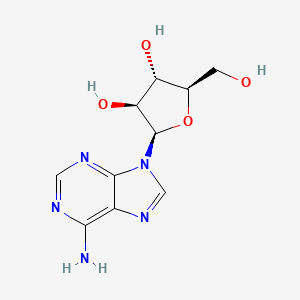 |
0.262 | ||
| ENC001032 | 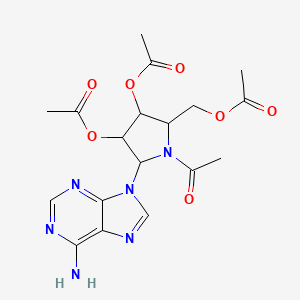 |
0.177 | D04KYY | 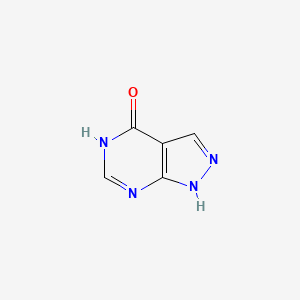 |
0.261 | ||
| ENC000577 | 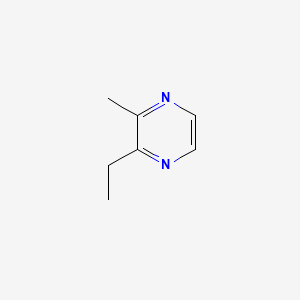 |
0.174 | D0RU0O | 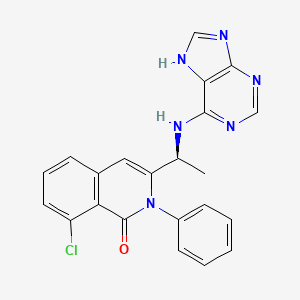 |
0.247 | ||