NPs Basic Information
|
Name |
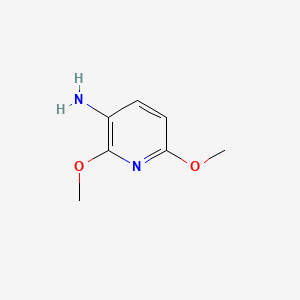
3-Amino-2,6-dimethoxypyridine
|
| Molecular Formula | C7H10N2O2 | |
| IUPAC Name* |
2,6-dimethoxypyridin-3-amine
|
|
| SMILES |
COC1=NC(=C(C=C1)N)OC
|
|
| InChI |
InChI=1S/C7H10N2O2/c1-10-6-4-3-5(8)7(9-6)11-2/h3-4H,8H2,1-2H3
|
|
| InChIKey |
PTBHRJOTANEONS-UHFFFAOYSA-N
|
|
| Synonyms |
3-Amino-2,6-dimethoxypyridine; 28020-37-3; 2,6-dimethoxypyridin-3-amine; 3-Pyridinamine, 2,6-dimethoxy-; 2,6-Dimethoxy-3-pyridinamine; Maybridge1_008017; SCHEMBL656848; 2,6-dimethoxypyridine-3-amine; 3-amino-2.6-dimethoxypyridine; HMS564E09; 2,6-dimethoxy-pyridin-3-ylamine; DTXSID40343903; 2,6-Dimethoxy-3-pyridinamine #; ZINC111531; MFCD00832865; AKOS009158431; CS-W005692; FS-2753; GK-0001; SB76443; 3-amino-2,6-dimethoxypyridine, AldrichCPR; DB-067864; FT-0647733; EN300-57156; 020A373; A876801; J-511528
|
|
| CAS | 28020-37-3 | |
| PubChem CID | 593005 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.17 | ALogp: | 0.8 |
| HBD: | 1 | HBA: | 4 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 57.4 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.692 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.463 | MDCK Permeability: | 0.00002390 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.066 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.002 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.835 | Plasma Protein Binding (PPB): | 64.40% |
| Volume Distribution (VD): | 0.938 | Fu: | 29.32% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.828 | CYP1A2-substrate: | 0.799 |
| CYP2C19-inhibitor: | 0.234 | CYP2C19-substrate: | 0.837 |
| CYP2C9-inhibitor: | 0.064 | CYP2C9-substrate: | 0.528 |
| CYP2D6-inhibitor: | 0.019 | CYP2D6-substrate: | 0.899 |
| CYP3A4-inhibitor: | 0.101 | CYP3A4-substrate: | 0.323 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.512 | Half-life (T1/2): | 0.31 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.055 | Human Hepatotoxicity (H-HT): | 0.305 |
| Drug-inuced Liver Injury (DILI): | 0.776 | AMES Toxicity: | 0.744 |
| Rat Oral Acute Toxicity: | 0.564 | Maximum Recommended Daily Dose: | 0.028 |
| Skin Sensitization: | 0.218 | Carcinogencity: | 0.256 |
| Eye Corrosion: | 0.014 | Eye Irritation: | 0.888 |
| Respiratory Toxicity: | 0.849 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000501 | 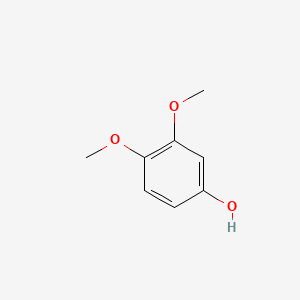 |
0.333 | D09GYT | 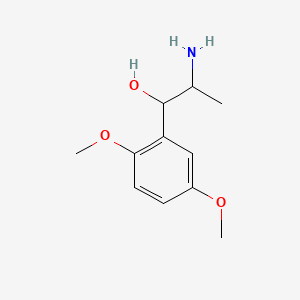 |
0.296 | ||
| ENC000168 | 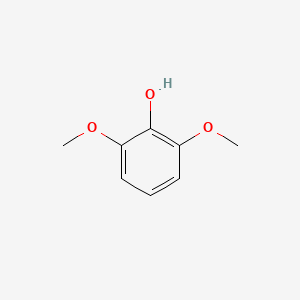 |
0.333 | D07PAO | 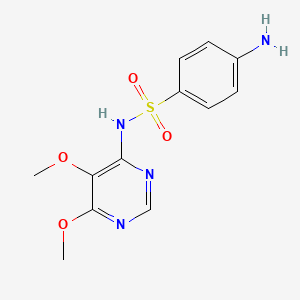 |
0.294 | ||
| ENC000712 | 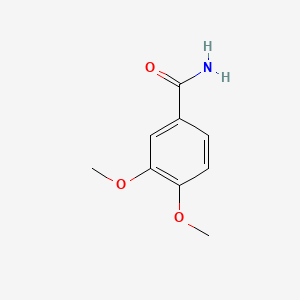 |
0.327 | D02XJY | 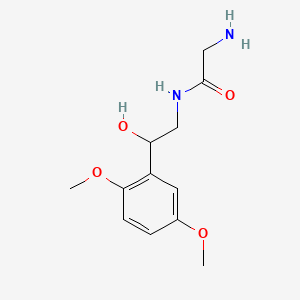 |
0.254 | ||
| ENC000478 | 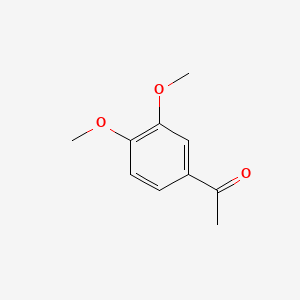 |
0.300 | D06QKV |  |
0.250 | ||
| ENC001461 | 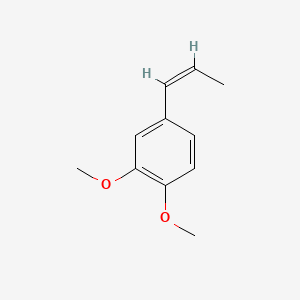 |
0.294 | D0AO5H | 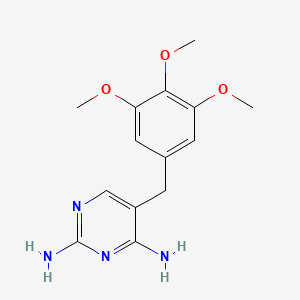 |
0.239 | ||
| ENC000499 | 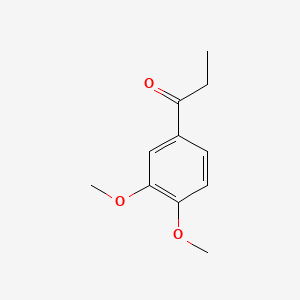 |
0.283 | D0G4YT | 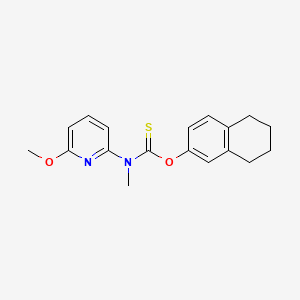 |
0.231 | ||
| ENC001363 | 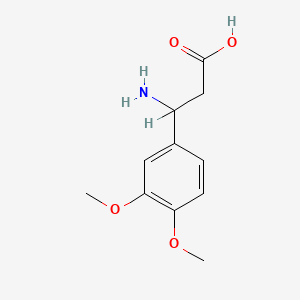 |
0.281 | D0E9CD | 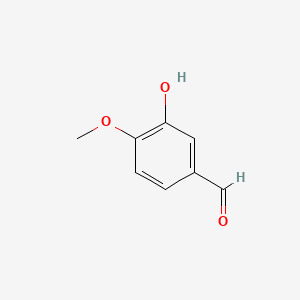 |
0.224 | ||
| ENC000736 | 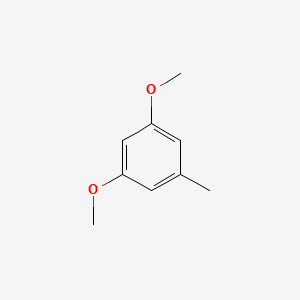 |
0.277 | D0Y7TS | 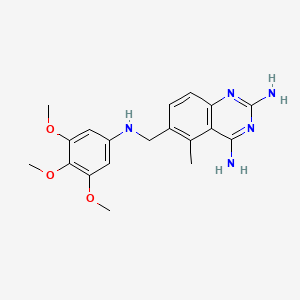 |
0.221 | ||
| ENC000349 | 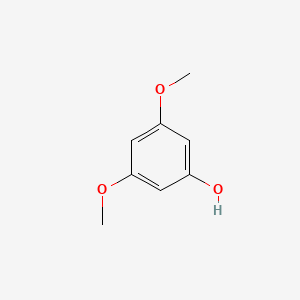 |
0.277 | D0Q4YI | 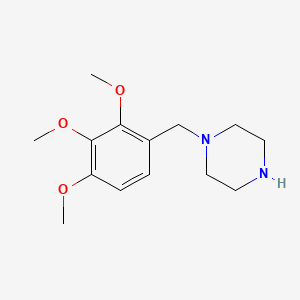 |
0.217 | ||
| ENC001512 | 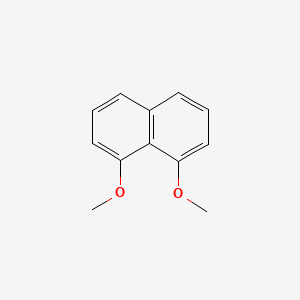 |
0.273 | D01XNB |  |
0.213 | ||