NPs Basic Information
|
Name |
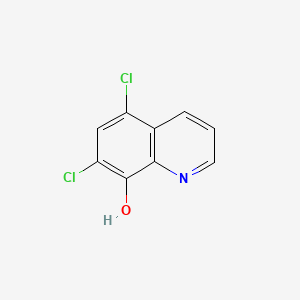
Chloroxine
|
| Molecular Formula | C9H5Cl2NO | |
| IUPAC Name* |
5,7-dichloroquinolin-8-ol
|
|
| SMILES |
C1=CC2=C(C(=C(C=C2Cl)Cl)O)N=C1
|
|
| InChI |
InChI=1S/C9H5Cl2NO/c10-6-4-7(11)9(13)8-5(6)2-1-3-12-8/h1-4,13H
|
|
| InChIKey |
WDFKMLRRRCGAKS-UHFFFAOYSA-N
|
|
| Synonyms |
chloroxine; 773-76-2; 5,7-Dichloroquinolin-8-ol; 5,7-Dichloro-8-hydroxyquinoline; 5,7-Dichloro-8-quinolinol; Capitrol; Chlorquinol; Dichloroxin; Quixalin; Chloroxyquinoline; Dichloroquinolinol; Dikhloroskin; Clofuzid; Endiaron; Quinolor; Dichlorohydroxyquinoline; Chlofucid; Quesyl; 5,7-Dichlorooxine; 5,7-Dichloroxine; 8-QUINOLINOL, 5,7-DICHLORO-; 5,7-Dichloro-8-oxyquinoline; Chlorohydroxyquinoline; Chloroxine [USAN]; 5,7-Dichlor-8-hydroxychinolin; NSC 3904; NSC-3904; MFCD00006786; 2I8BD50I8B; 5,7-Dichloro-8-hydroquinoline; CHEBI:59477; NSC3904; Chloroxine (USAN); AS-229; NCGC00095264-01; DSSTox_CID_2801; DSSTox_RID_76734; DSSTox_GSID_22801; CAS-773-76-2; CCRIS 5751; Capitrol Cream Shampoo; SR-01000747474; EINECS 212-258-3; BRN 0153606; UNII-2I8BD50I8B; 5,7-Dichlor-8-hydroxychinolin [German]; SQ 16401; Capitrol (TN); Spectrum_001434; CHLOROXINE [MI]; 5,7-Dichloro-8-oxine; Spectrum2_000687; Spectrum3_001156; Spectrum4_000744; Spectrum5_001444; CHLOROXINE [VANDF]; 8-Quinolinol,7-dichloro-; CHLOROXINE [MART.]; cid_2722; SCHEMBL3350; 5,7-Dichloro-8-Quinoliol; CHLOROXINE [WHO-DD]; Oprea1_486275; REGID_for_CID_2722; BSPBio_002711; KBioGR_001068; KBioSS_001914; 5-21-03-00286 (Beilstein Handbook Reference); MLS000681736; BIDD:GT0763; DivK1c_000578; SPECTRUM1503202; SPBio_000813; ZINC1131; CHEMBL1200596; CHLOROXINE [ORANGE BOOK]; DTXSID5022801; BDBM32147; HMS501M20; KBio1_000578; KBio2_001914; KBio2_004482; KBio2_007050; KBio3_002211; NINDS_000578; HMS1923K05; HMS2092N04; HMS2609A16; HMS3259E17; HMS3655G16; HMS3712O06; Pharmakon1600-01503202; Capitrol Cream Shampoo (Salt/Mix); AMY23282; component of Capitrol Cream Shampoo; HY-B0295; 5,7-Dichloro-8-quinolinol, 99%; Tox21_111494; AC9195; CCG-40039; NSC758398; s1839; STK075368; 5,7-dichloroquinolin-8-ol;chloroxine; AKOS000271338; Tox21_111494_1; AC-4820; DB01243; NC00505; NSC-758398; IDI1_000578; UPCMLD0ENAT5752182:001; NCGC00095264-02; NCGC00095264-03; NCGC00095264-04; NCGC00095264-05; SMR000312779; SY016017; TG2-36-1; SBI-0051782.P002; A9795; D0412; FT-0623709; SW198618-2; D03472; EN300-207135; AB00052323-08; AB00052323_09; AB00052323_10; SR-01000747474-2; SR-01000747474-3; W-104320; BRD-K17075857-001-06-9; Q12029435; F0918-0943; Z111781330
|
|
| CAS | 773-76-2 | |
| PubChem CID | 2722 | |
| ChEMBL ID | CHEMBL1200596 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 214.04 | ALogp: | 3.5 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 33.1 | Aromatic Rings: | 2 |
| Heavy Atoms: | 13 | QED Weighted: | 0.718 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.639 | MDCK Permeability: | 0.00002570 |
| Pgp-inhibitor: | 0.007 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.031 |
| 30% Bioavailability (F30%): | 0.98 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.26 | Plasma Protein Binding (PPB): | 97.84% |
| Volume Distribution (VD): | 1.15 | Fu: | 1.64% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.958 | CYP1A2-substrate: | 0.744 |
| CYP2C19-inhibitor: | 0.309 | CYP2C19-substrate: | 0.468 |
| CYP2C9-inhibitor: | 0.331 | CYP2C9-substrate: | 0.863 |
| CYP2D6-inhibitor: | 0.214 | CYP2D6-substrate: | 0.619 |
| CYP3A4-inhibitor: | 0.078 | CYP3A4-substrate: | 0.16 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.863 | Half-life (T1/2): | 0.363 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.027 | Human Hepatotoxicity (H-HT): | 0.083 |
| Drug-inuced Liver Injury (DILI): | 0.945 | AMES Toxicity: | 0.187 |
| Rat Oral Acute Toxicity: | 0.26 | Maximum Recommended Daily Dose: | 0.19 |
| Skin Sensitization: | 0.872 | Carcinogencity: | 0.507 |
| Eye Corrosion: | 0.009 | Eye Irritation: | 0.786 |
| Respiratory Toxicity: | 0.94 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000117 |  |
0.277 | D02HWP |  |
1.000 | ||
| ENC002806 |  |
0.273 | D06AEB | 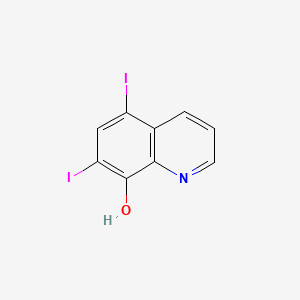 |
0.532 | ||
| ENC004703 | 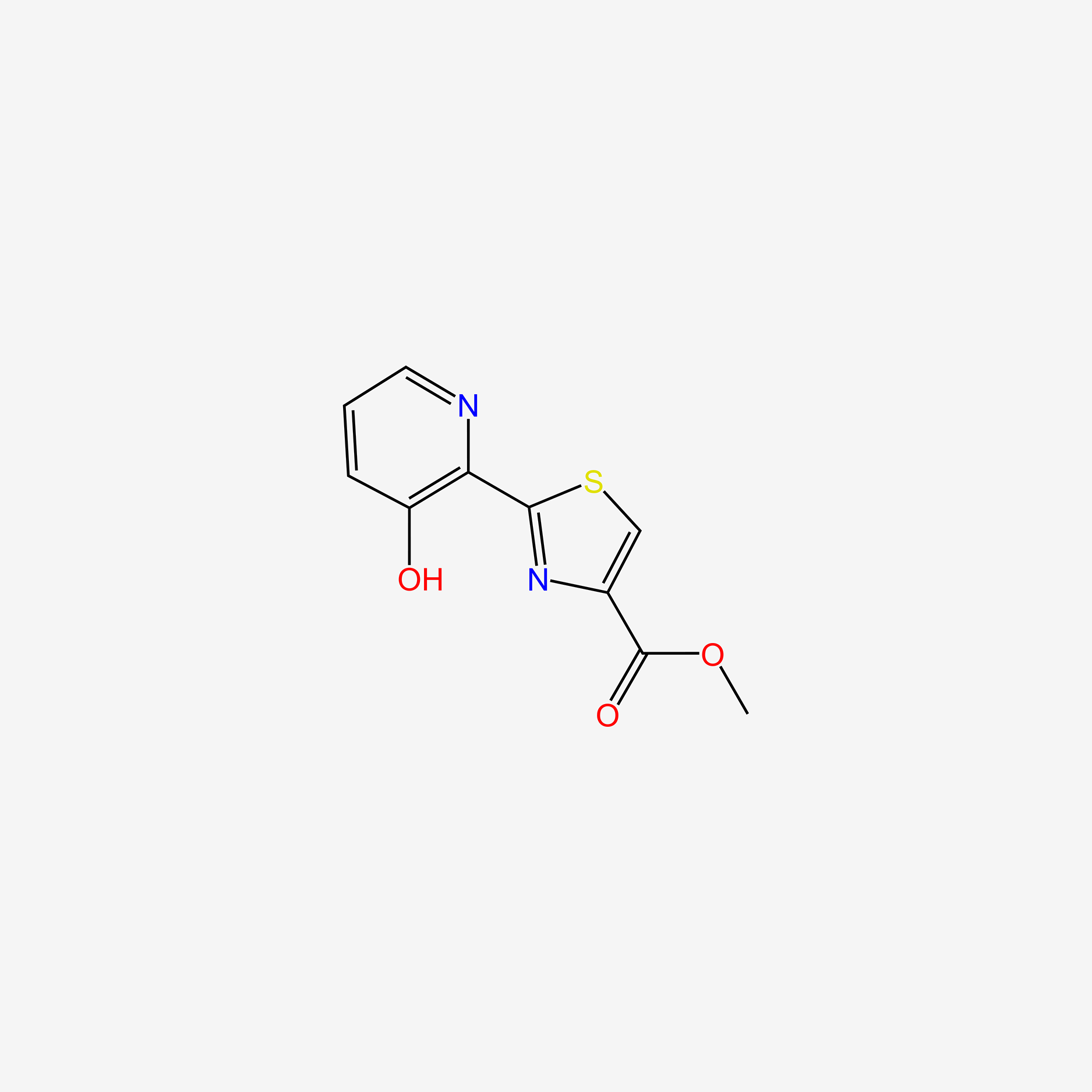 |
0.266 | D09SOA | 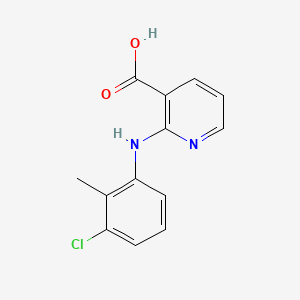 |
0.303 | ||
| ENC005704 | 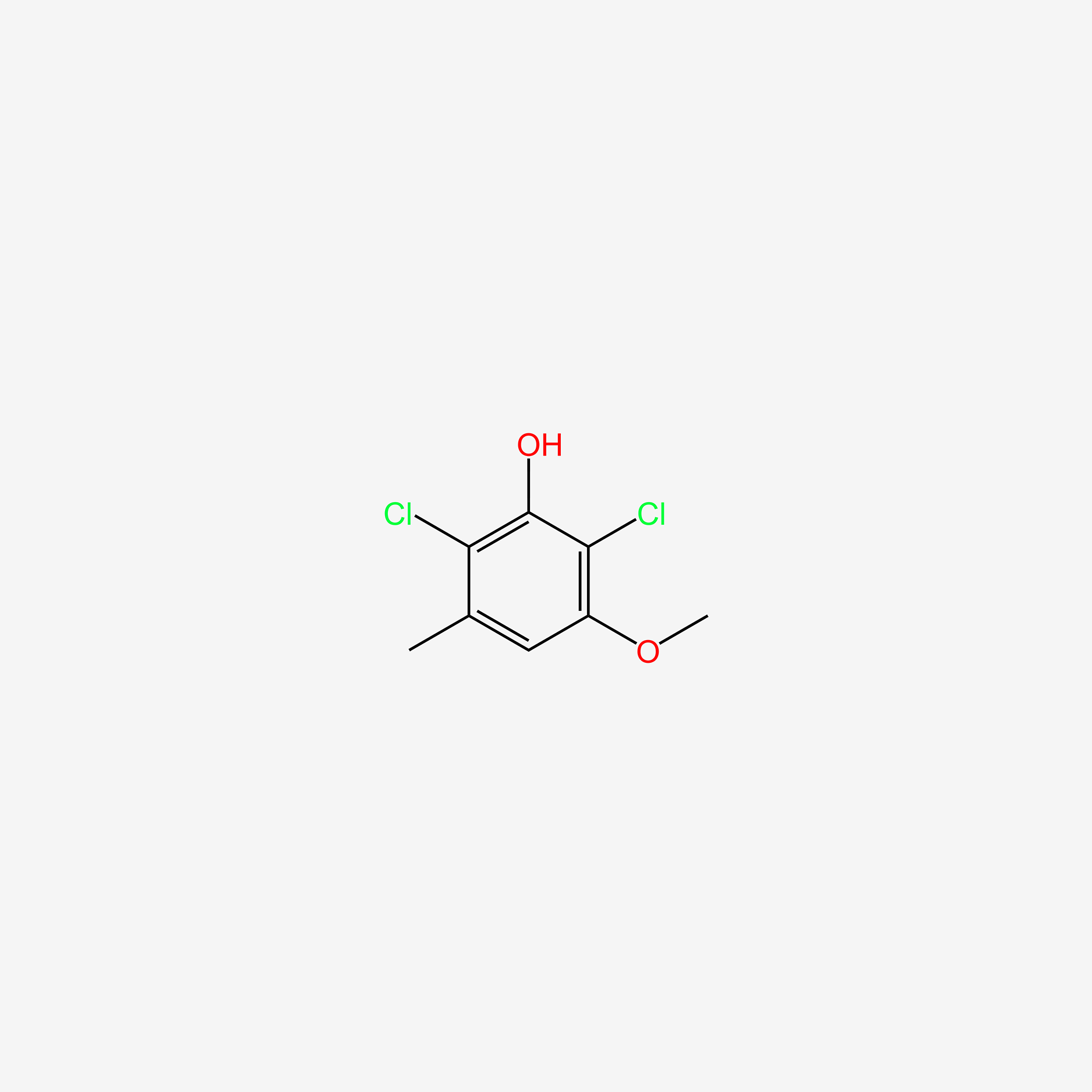 |
0.264 | D00CSQ | 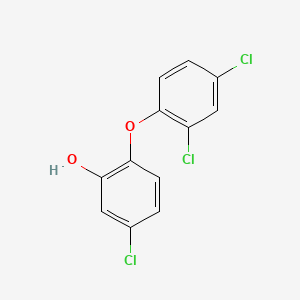 |
0.277 | ||
| ENC002809 | 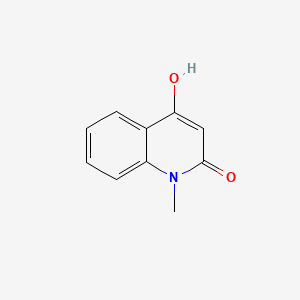 |
0.263 | D01IEM | 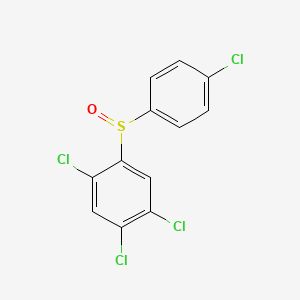 |
0.269 | ||
| ENC000118 | 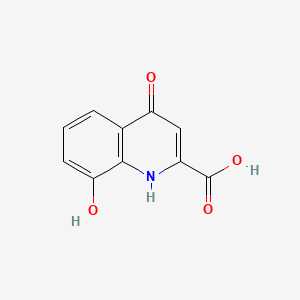 |
0.262 | D03SKR | 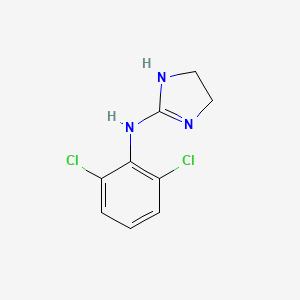 |
0.267 | ||
| ENC005702 | 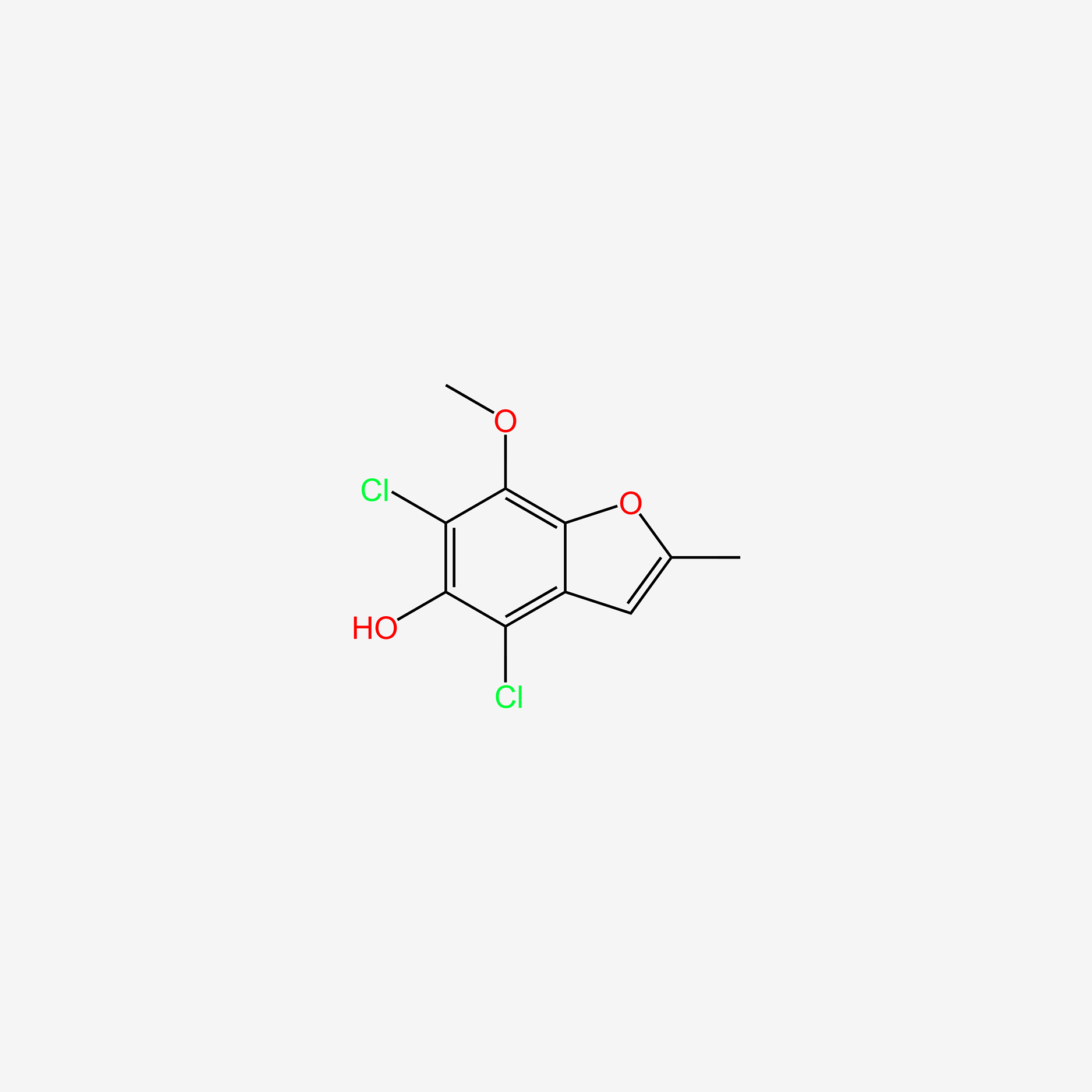 |
0.246 | D0ZX2G | 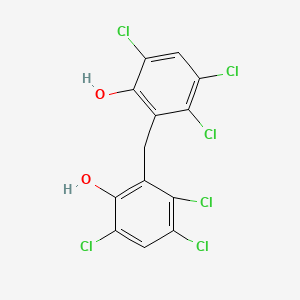 |
0.264 | ||
| ENC000056 | 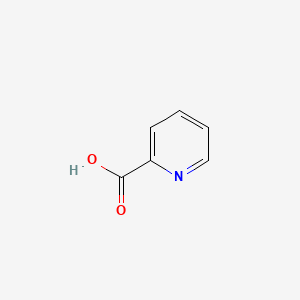 |
0.245 | D0TG1H | 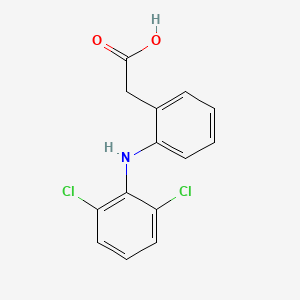 |
0.254 | ||
| ENC001978 | 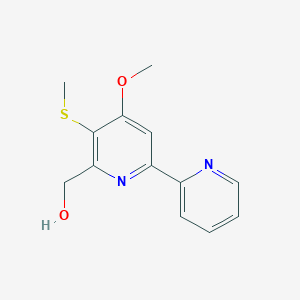 |
0.243 | D0T1LK | 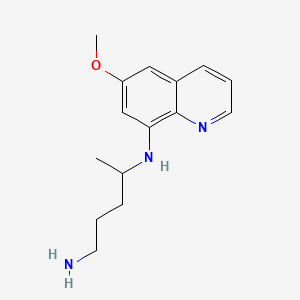 |
0.250 | ||
| ENC005053 | 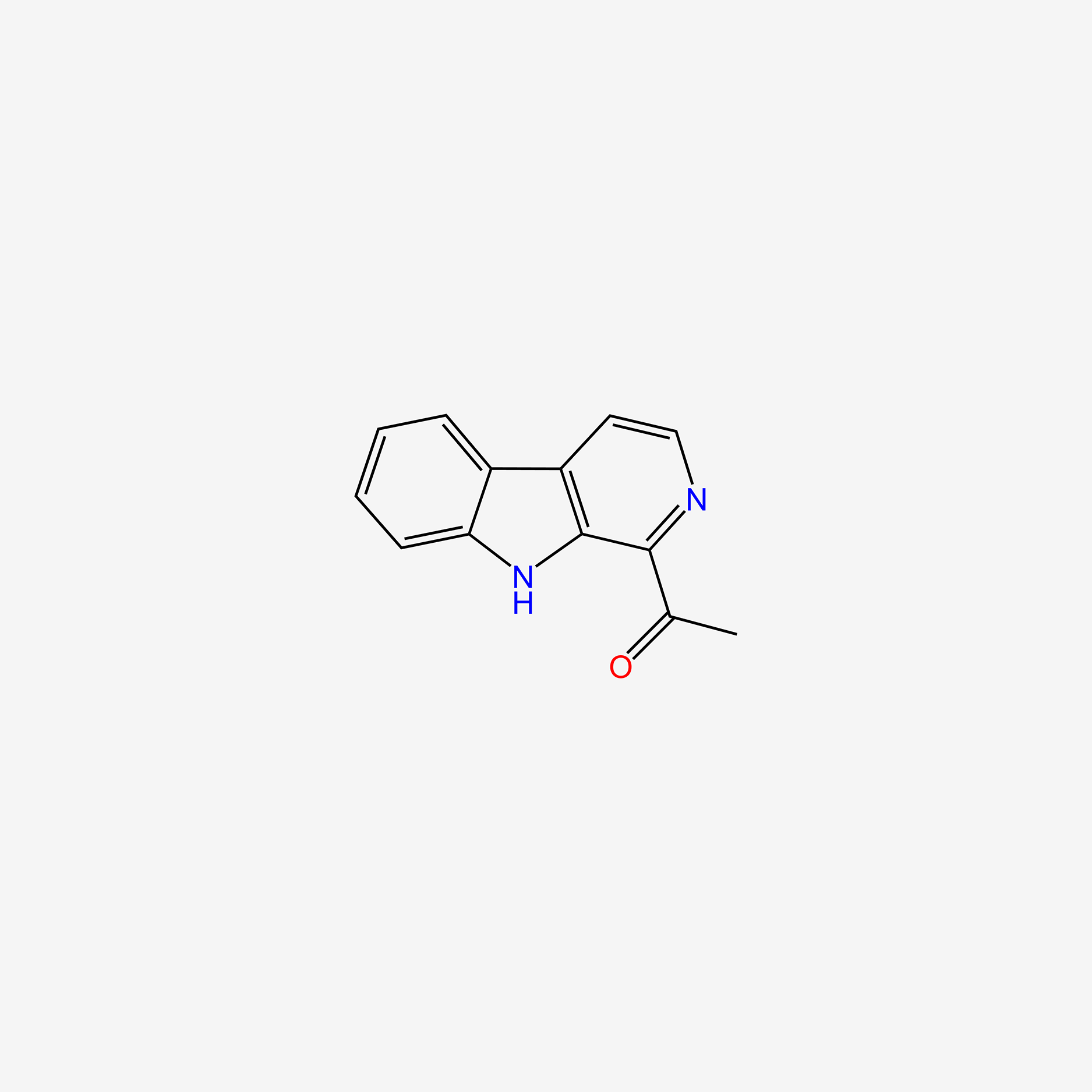 |
0.242 | D0W6KM |  |
0.250 | ||