NPs Basic Information
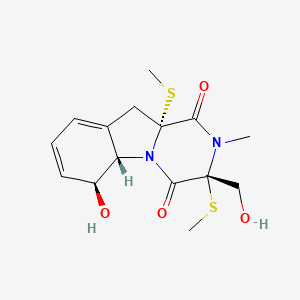
|
Name |
Bisdethiobis(methylthio)gliotoxin
|
| Molecular Formula | C15H20N2O4S2 | |
| IUPAC Name* |
(3R,5aS,6S,10aR)-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylsulfanyl)-6,10-dihydro-5aH-pyrazino[1,2-a]indole-1,4-dione
|
|
| SMILES |
CN1C(=O)[C@@]2(CC3=CC=C[C@@H]([C@H]3N2C(=O)[C@@]1(CO)SC)O)SC
|
|
| InChI |
InChI=1S/C15H20N2O4S2/c1-16-12(20)14(22-2)7-9-5-4-6-10(19)11(9)17(14)13(21)15(16,8-18)23-3/h4-6,10-11,18-19H,7-8H2,1-3H3/t10-,11-,14+,15+/m0/s1
|
|
| InChIKey |
OVBAGMZLGLXSBN-UOVKNHIHSA-N
|
|
| Synonyms |
Bisdethiobis(methylthio)gliotoxin; bis(methylthio)gliotoxin; 74149-38-5; FR-49175; FR 49175; (3R,5aS,6S,10aR)-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylsulfanyl)-6,10-dihydro-5aH-pyrazino[1,2-a]indole-1,4-dione; CHEMBL1088981; dimethylgliotoxin; AC1L4YI4; BmGT; S,S-Dimethyl gliotoxin; bis(methylsulfanyl)gliotoxin; Bis(methylthio)gliotoxin, 99%; GTPL3150; SCHEMBL18652994; DTXSID70995590; 3R,10Ar-Dithiomethyl Gliotoxin A; HY-N9710; ZINC5599818; BDBM50315535; MFCD00133146; Bis(methylthio)gliotoxin (FR-49175); NCGC00380667-01; CS-0203642; Q27077740; Bis(methylthio)gliotoxin (FR-49175) is known as a PAF receptor ligand.; (3R,5aS,6S,10aR)-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylsulfanyl)-1H,2H,3H,4H,5aH,6H,10H,10aH-piperazino[1,2-a]indole-1,4-dione; (3R,5aS,6S,10aR)-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylthio)-2,3,5a,6,10,10a-hexahydropyrazino[1,2-a]indole-1,4-dione; 6-Hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylsulfanyl)-2,3,5a,6,10,10a-hexahydropyrazino[1,2-a]indole-1,4-dione; NCGC00380667-01_C15H20N2O4S2_Pyrazino[1,2-a]indole-1,4-dione, 2,3,5a,6,10,10a-hexahydro-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylthio)-, (3R,5aS,6S,10aR)-; Pyrazino(1,2-a)indole-1,4-dione, 2,3,5a,6,10,10a-hexahydro-6-hydroxy-3-(hydroxymethyl)-2-methyl-3,10a-bis(methylthio)-, (3R-(3alpha,5abeta,6beta,10aalpha))-
|
|
| CAS | 74149-38-5 | |
| PubChem CID | 194564 | |
| ChEMBL ID | CHEMBL1088981 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 356.5 | ALogp: | 0.1 |
| HBD: | 2 | HBA: | 6 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 132.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 23 | QED Weighted: | 0.763 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.001 | MDCK Permeability: | 0.00002020 |
| Pgp-inhibitor: | 0.009 | Pgp-substrate: | 0.042 |
| Human Intestinal Absorption (HIA): | 0.282 | 20% Bioavailability (F20%): | 0.006 |
| 30% Bioavailability (F30%): | 0.004 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.786 | Plasma Protein Binding (PPB): | 81.35% |
| Volume Distribution (VD): | 0.658 | Fu: | 34.35% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.007 | CYP1A2-substrate: | 0.106 |
| CYP2C19-inhibitor: | 0.157 | CYP2C19-substrate: | 0.912 |
| CYP2C9-inhibitor: | 0.604 | CYP2C9-substrate: | 0.079 |
| CYP2D6-inhibitor: | 0.002 | CYP2D6-substrate: | 0.067 |
| CYP3A4-inhibitor: | 0.087 | CYP3A4-substrate: | 0.97 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.132 | Half-life (T1/2): | 0.901 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.19 |
| Drug-inuced Liver Injury (DILI): | 0.965 | AMES Toxicity: | 0.024 |
| Rat Oral Acute Toxicity: | 0.6 | Maximum Recommended Daily Dose: | 0.736 |
| Skin Sensitization: | 0.834 | Carcinogencity: | 0.846 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.008 |
| Respiratory Toxicity: | 0.074 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC006009 | 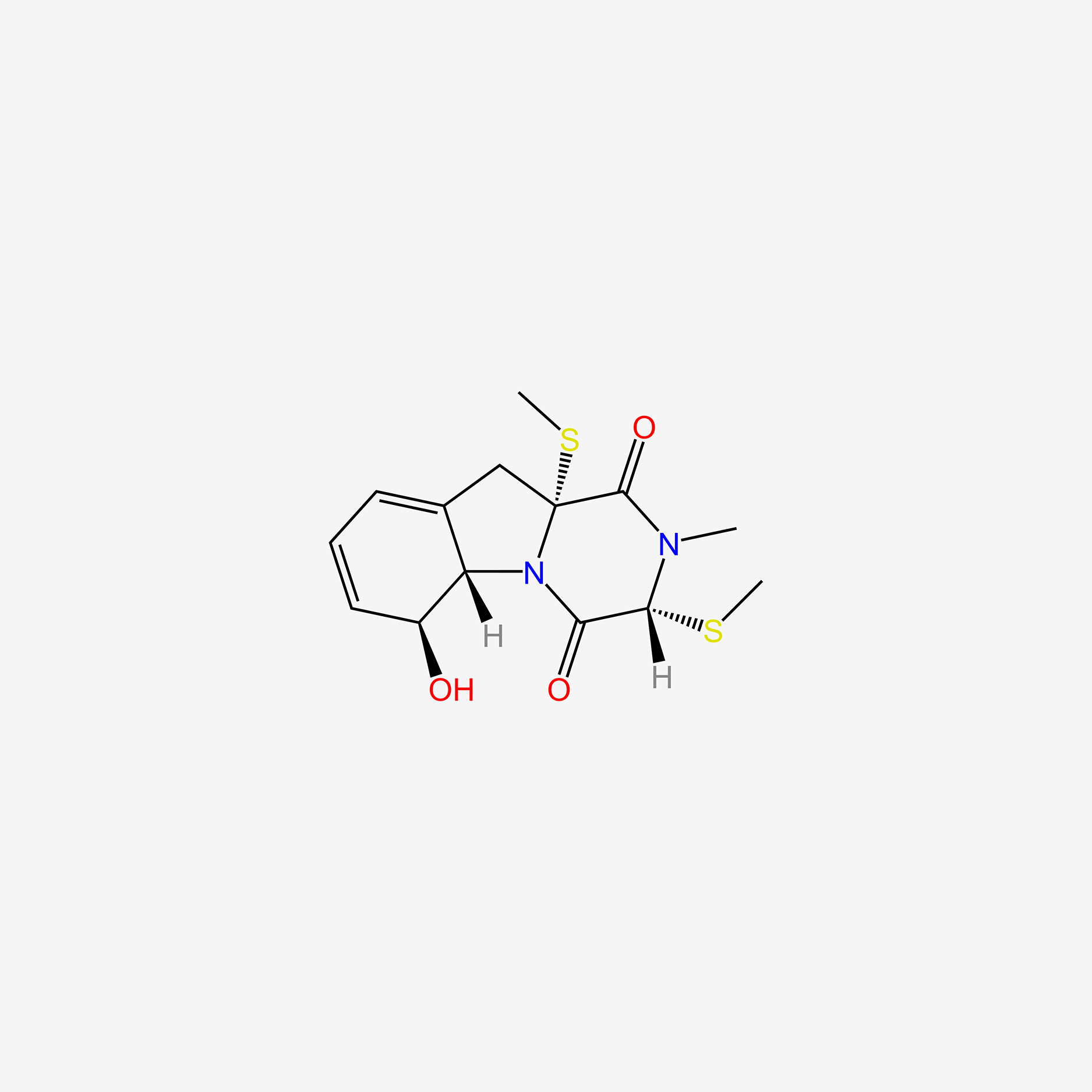 |
0.630 | D08EOD |  |
0.212 | ||
| ENC005509 | 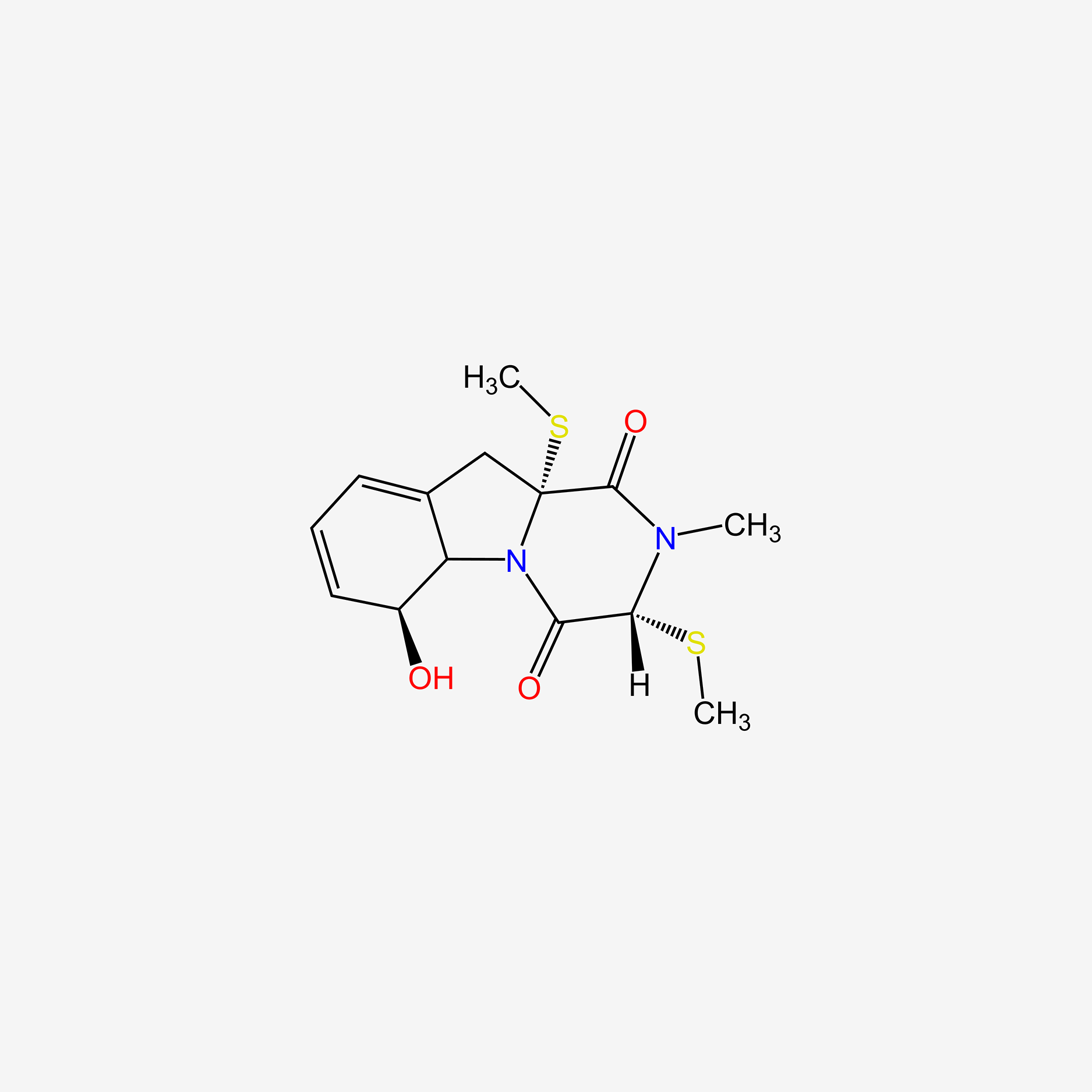 |
0.630 | D0I5DS | 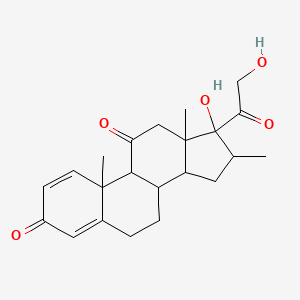 |
0.205 | ||
| ENC000134 | 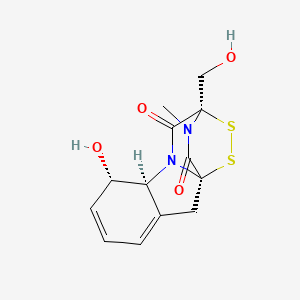 |
0.538 | D0Z8EX | 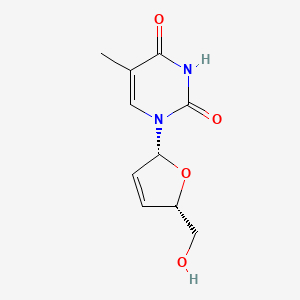 |
0.205 | ||
| ENC005510 | 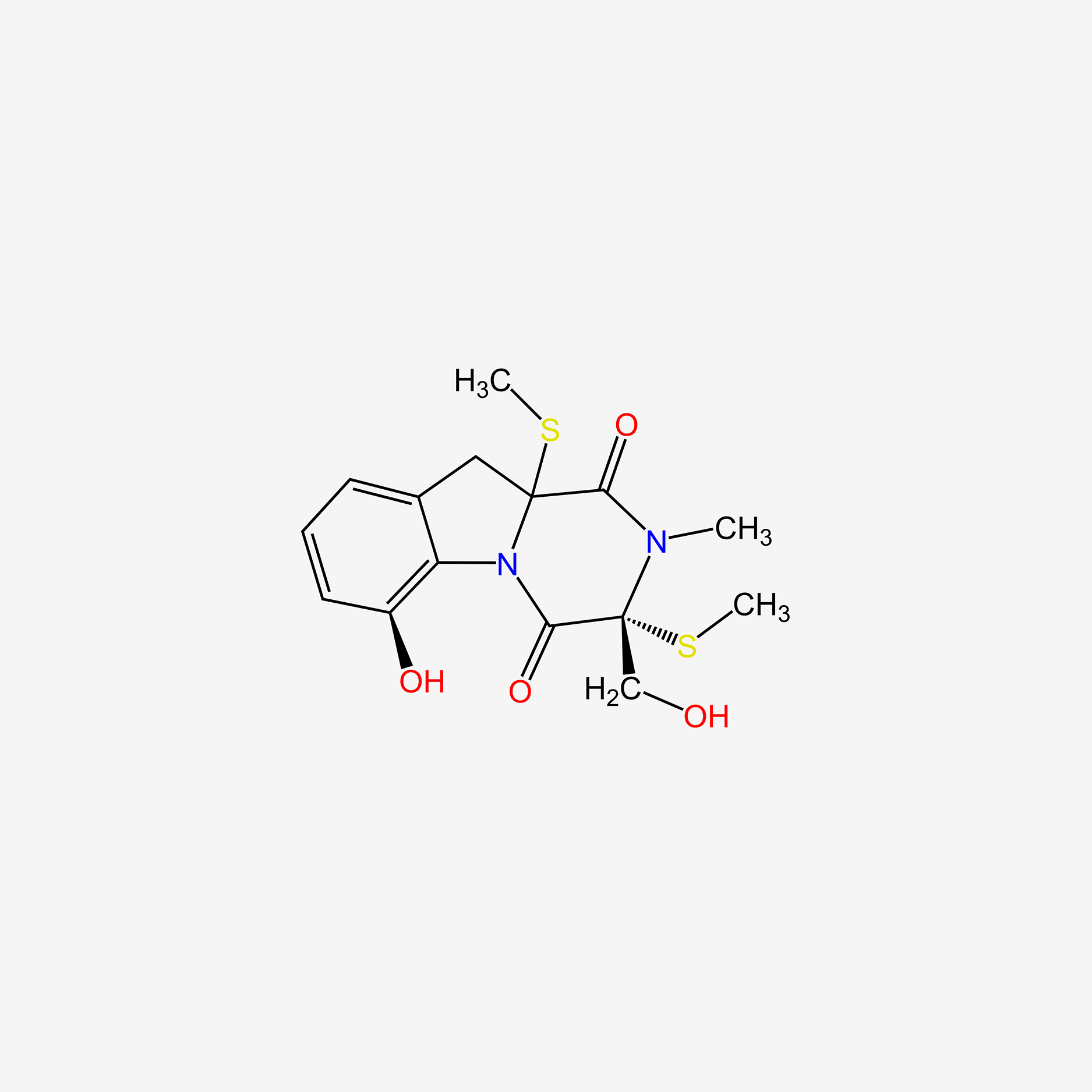 |
0.512 | D0G6AB |  |
0.204 | ||
| ENC003617 |  |
0.442 | D0R2KF |  |
0.202 | ||
| ENC003438 |  |
0.432 | D0CL9S |  |
0.200 | ||
| ENC003035 | 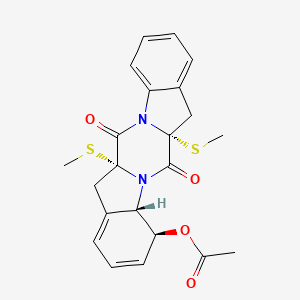 |
0.417 | D0IL7L | 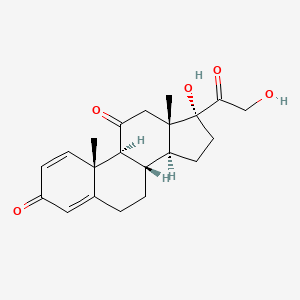 |
0.198 | ||
| ENC003595 | 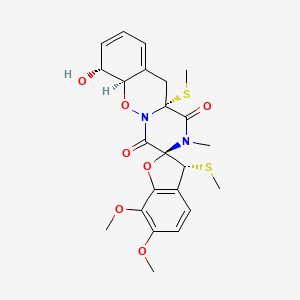 |
0.393 | D0W7RJ | 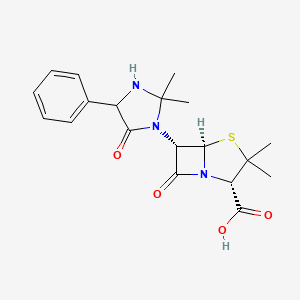 |
0.195 | ||
| ENC004752 |  |
0.353 | D07RGW |  |
0.191 | ||
| ENC003549 |  |
0.314 | D0TS1Z |  |
0.187 | ||