NPs Basic Information
|
Name |
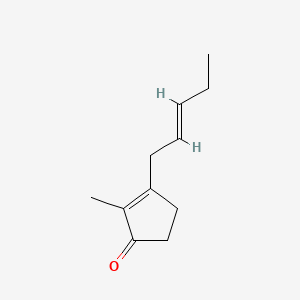
Isojasmone
|
| Molecular Formula | C11H16O | |
| IUPAC Name* |
2-methyl-3-[(E)-pent-2-enyl]cyclopent-2-en-1-one
|
|
| SMILES |
CC/C=C/CC1=C(C(=O)CC1)C
|
|
| InChI |
InChI=1S/C11H16O/c1-3-4-5-6-10-7-8-11(12)9(10)2/h4-5H,3,6-8H2,1-2H3/b5-4+
|
|
| InChIKey |
GVONPEQEUQYVNH-SNAWJCMRSA-N
|
|
| Synonyms |
Isojasmone; 11050-62-7; 2-Methyl-3-(2-pentenyl)-2-cyclopenten-1-one; 2-Cyclopenten-1-one, 2-methyl-3-(2-pentenyl)-; 2-methyl-3-[(E)-pent-2-enyl]cyclopent-2-en-1-one; TY8GSG1D56; 2-Methyl-3-pent-2-enylcyclopent-2-enone; 2-Cyclopenten-1-one, 2-methyl-3-(2-penten-1-yl)-; Isojasmone (natural); 2-Methyl-3-(2-penten-1-yl)-2-Cyclopenten-1-one; EINECS 234-273-4; 2-METHYL-3-((E)-PENT-2-ENYL)CYCLOPENT-2-EN-1-ONE; UNII-TY8GSG1D56; SCHEMBL1235273; CHEBI:189993; 152982-57-5
|
|
| CAS | 11050-62-7 | |
| PubChem CID | 6435841 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 164.24 | ALogp: | 2.2 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 12 | QED Weighted: | 0.58 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.466 | MDCK Permeability: | 0.00002470 |
| Pgp-inhibitor: | 0.474 | Pgp-substrate: | 0.004 |
| Human Intestinal Absorption (HIA): | 0.007 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.002 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.972 | Plasma Protein Binding (PPB): | 90.82% |
| Volume Distribution (VD): | 1.076 | Fu: | 8.23% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.115 | CYP1A2-substrate: | 0.396 |
| CYP2C19-inhibitor: | 0.049 | CYP2C19-substrate: | 0.889 |
| CYP2C9-inhibitor: | 0.035 | CYP2C9-substrate: | 0.599 |
| CYP2D6-inhibitor: | 0.054 | CYP2D6-substrate: | 0.884 |
| CYP3A4-inhibitor: | 0.033 | CYP3A4-substrate: | 0.367 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.592 | Half-life (T1/2): | 0.765 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.018 | Human Hepatotoxicity (H-HT): | 0.145 |
| Drug-inuced Liver Injury (DILI): | 0.078 | AMES Toxicity: | 0.109 |
| Rat Oral Acute Toxicity: | 0.594 | Maximum Recommended Daily Dose: | 0.845 |
| Skin Sensitization: | 0.823 | Carcinogencity: | 0.893 |
| Eye Corrosion: | 0.182 | Eye Irritation: | 0.857 |
| Respiratory Toxicity: | 0.714 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001459 | 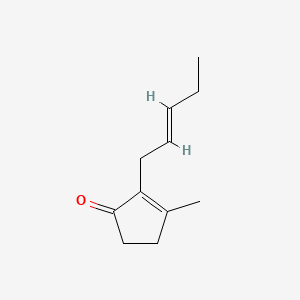 |
0.650 | D0H6VY |  |
0.190 | ||
| ENC000476 | 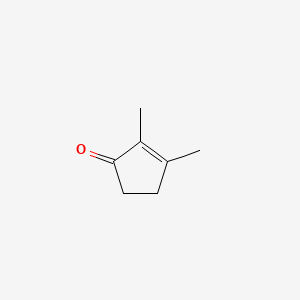 |
0.421 | D09TPF | 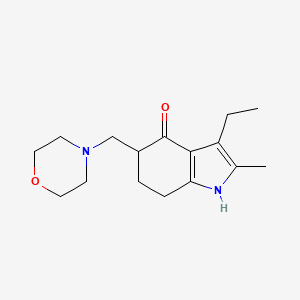 |
0.184 | ||
| ENC002343 |  |
0.326 | D00IUG |  |
0.182 | ||
| ENC005598 |  |
0.305 | D0Q4XQ |  |
0.180 | ||
| ENC001581 | 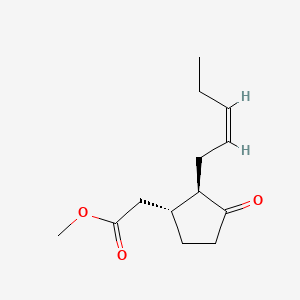 |
0.305 | D05OQJ | 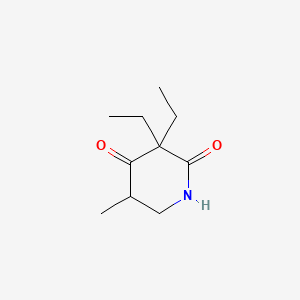 |
0.175 | ||
| ENC001746 | 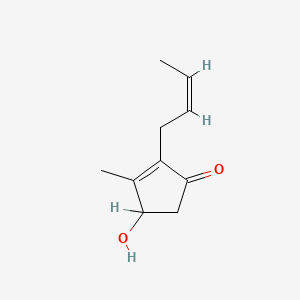 |
0.300 | D0E0WQ | 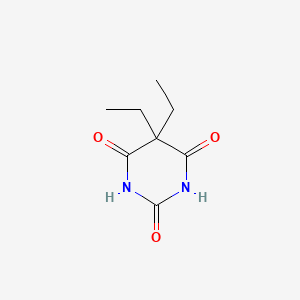 |
0.175 | ||
| ENC002751 | 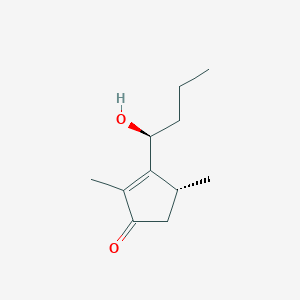 |
0.264 | D0W0MF | 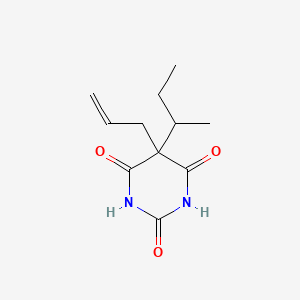 |
0.172 | ||
| ENC005984 | 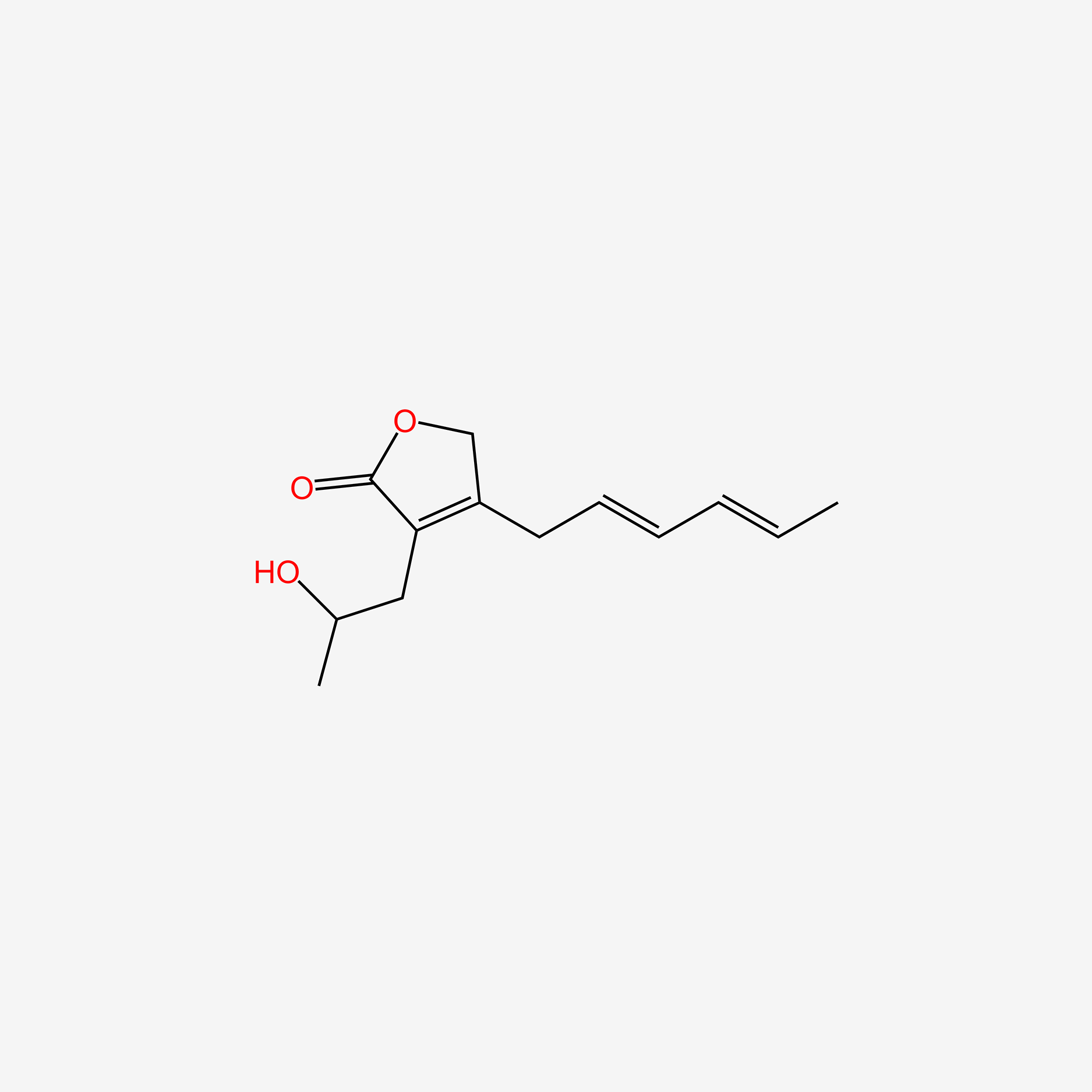 |
0.262 | D02OZY | 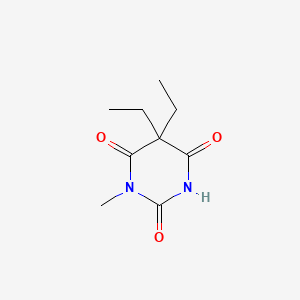 |
0.169 | ||
| ENC006016 | 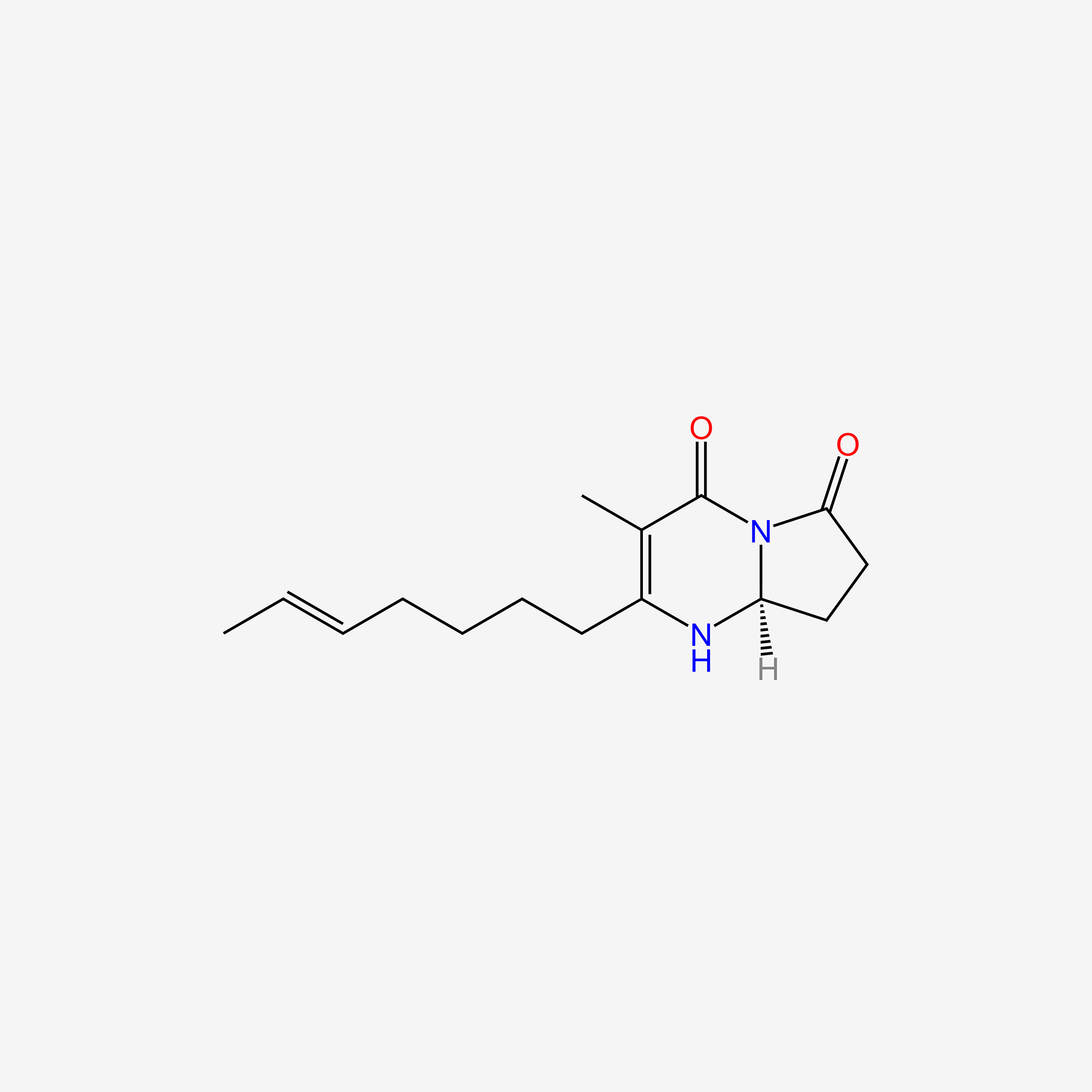 |
0.246 | D03GCJ | 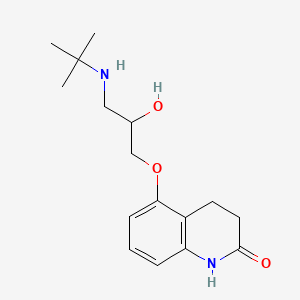 |
0.167 | ||
| ENC004598 | 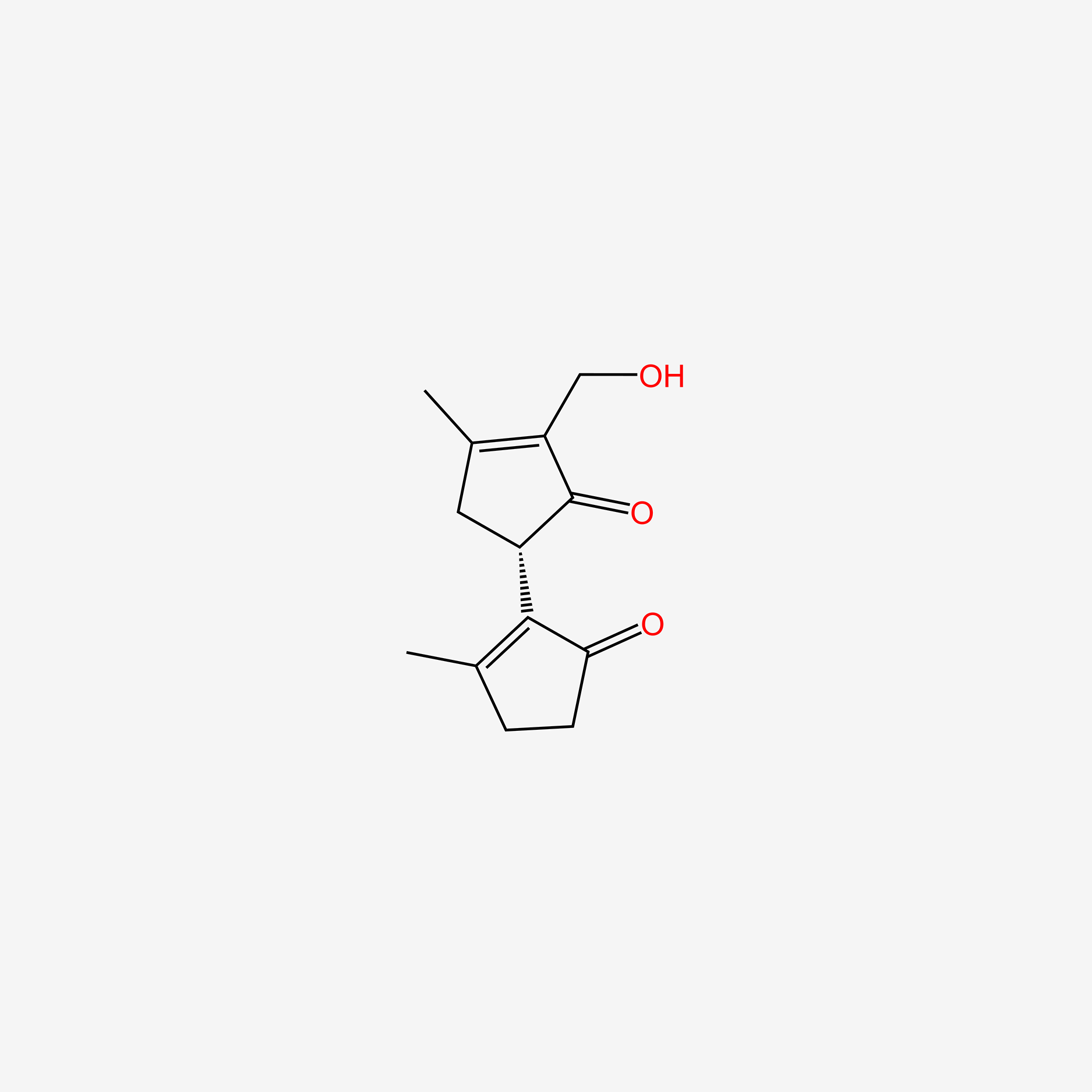 |
0.246 | D00MIN | 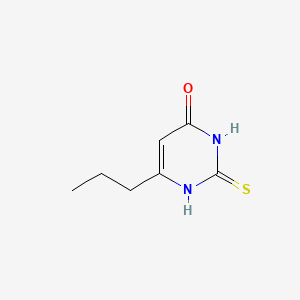 |
0.167 | ||