NPs Basic Information
|
Name |
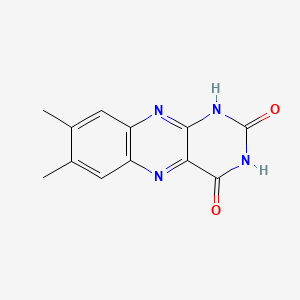
Lumichrome
|
| Molecular Formula | C12H10N4O2 | |
| IUPAC Name* |
7,8-dimethyl-1H-benzo[g]pteridine-2,4-dione
|
|
| SMILES |
CC1=CC2=C(C=C1C)N=C3C(=N2)C(=O)NC(=O)N3
|
|
| InChI |
InChI=1S/C12H10N4O2/c1-5-3-7-8(4-6(5)2)14-10-9(13-7)11(17)16-12(18)15-10/h3-4H,1-2H3,(H2,14,15,16,17,18)
|
|
| InChIKey |
ZJTJUVIJVLLGSP-UHFFFAOYSA-N
|
|
| Synonyms |
lumichrome; 7,8-Dimethylalloxazine; 1086-80-2; 7,8-dimethylbenzo[g]pteridine-2,4(1H,3H)-dione; Riboflavin lumichrome; Alloxazine, 7,8-dimethyl-; 7,8-dimethylisoalloxazine; Benzo[g]pteridine-2,4(1H,3H)-dione, 7,8-dimethyl-; Benzo(g)pteridine-2,4(1H,3H)-dione, 7,8-dimethyl-; 99U1UDJ2HM; CHEBI:17781; 7,8-dimethyl-1H-benzo[g]pteridine-2,4-dione; NSC-96911; 7,8-dimethylbenzo[g]pteridine-2,4(3H,10H)-dione; LUM; 7,8-dimethyl-10H-benzo[g]pteridine-2,4-dione; 7,8-Dimethylbenzo(g)pteridine-2,4(1H,3H)-dione; UNII-99U1UDJ2HM; Lumichrome (I); dimethylisoalloxazine; EINECS 214-120-8; MFCD00005021; NSC 96911; 2cc7; Alloxazine,8-dimethyl-; LUMICHROME [MI]; Oprea1_454036; SCHEMBL194906; CHEMBL1234101; CHEBI:37323; DTXSID70148600; HMS3604B12; ALBB-023287; NSC96911; Riboflavin Impurity 2 (Lumichrome); CCG-36441; ZINC12446789; AKOS003232003; AKOS003245099; DB04345; AS-56222; 7,8-Dimethylbenzo[g]pteridine-2,4-diol #; DB-040839; HY-115385; RIBOFLAVIN IMPURITY B [EP IMPURITY]; CS-0081855; D1066; FT-0603542; C01727; D78457; 7,8-dimethyl-1H-benzo[g]pteridine-2,4-quinone; Benzo[g]pteridine-2,3H)-dione, 7,8-dimethyl-; SR-01000198213; J-002180; SR-01000198213-1; Q27095157; 3CC7321C-61C5-4305-A0F0-EE1334A0C8A1; 7,8-DIMETHYLBENZO(G)PTERIDINE-2,4-(1H,3H)-DIONE; 7,8-Dimethylbenzo[g]pteridine-2,4(1H,3H)-dione (Lumichrome)
|
|
| CAS | 1086-80-2 | |
| PubChem CID | 5326566 | |
| ChEMBL ID | CHEMBL1234101 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 242.23 | ALogp: | 1.1 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 84.0 | Aromatic Rings: | 3 |
| Heavy Atoms: | 18 | QED Weighted: | 0.579 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.926 | MDCK Permeability: | 0.00001230 |
| Pgp-inhibitor: | 0.003 | Pgp-substrate: | 0.094 |
| Human Intestinal Absorption (HIA): | 0.572 | 20% Bioavailability (F20%): | 0.012 |
| 30% Bioavailability (F30%): | 0.003 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.042 | Plasma Protein Binding (PPB): | 94.56% |
| Volume Distribution (VD): | 0.407 | Fu: | 4.04% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.956 | CYP1A2-substrate: | 0.986 |
| CYP2C19-inhibitor: | 0.069 | CYP2C19-substrate: | 0.058 |
| CYP2C9-inhibitor: | 0.01 | CYP2C9-substrate: | 0.032 |
| CYP2D6-inhibitor: | 0.017 | CYP2D6-substrate: | 0.035 |
| CYP3A4-inhibitor: | 0.038 | CYP3A4-substrate: | 0.258 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.674 | Half-life (T1/2): | 0.835 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.073 | Human Hepatotoxicity (H-HT): | 0.388 |
| Drug-inuced Liver Injury (DILI): | 0.987 | AMES Toxicity: | 0.54 |
| Rat Oral Acute Toxicity: | 0.333 | Maximum Recommended Daily Dose: | 0.716 |
| Skin Sensitization: | 0.061 | Carcinogencity: | 0.649 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.094 |
| Respiratory Toxicity: | 0.979 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001115 | 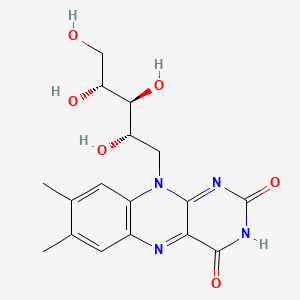 |
0.367 | D04QST | 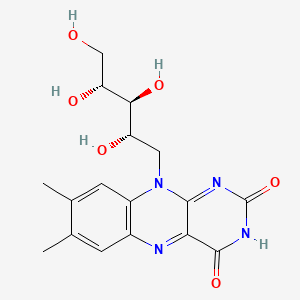 |
0.367 | ||
| ENC000063 | 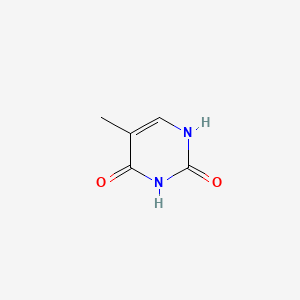 |
0.298 | D01PZD | 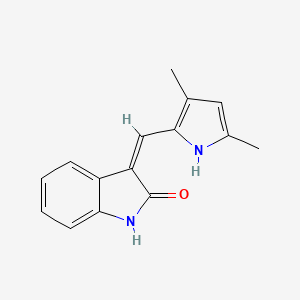 |
0.247 | ||
| ENC000181 | 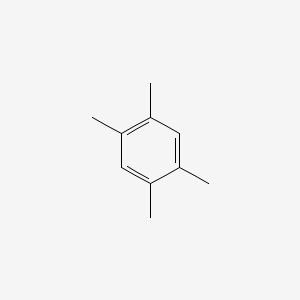 |
0.267 | D0C6DT | 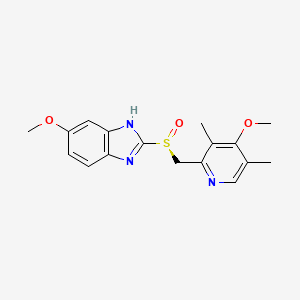 |
0.245 | ||
| ENC002723 | 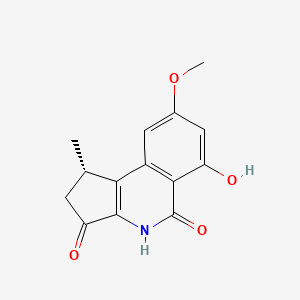 |
0.259 | D01XNB |  |
0.245 | ||
| ENC000066 | 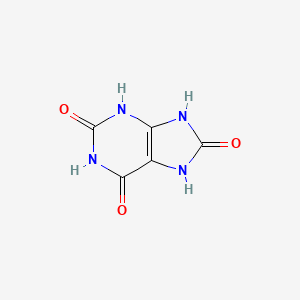 |
0.258 | D0E0SW | 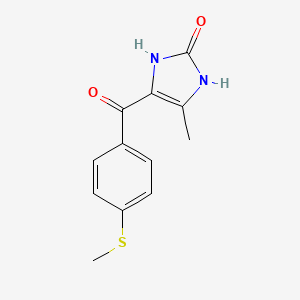 |
0.244 | ||
| ENC001026 | 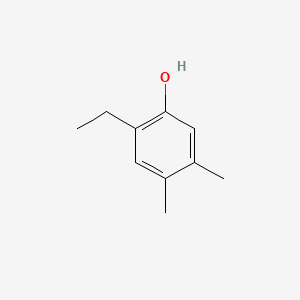 |
0.254 | D0FA2O | 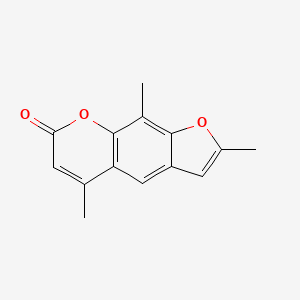 |
0.244 | ||
| ENC005958 | 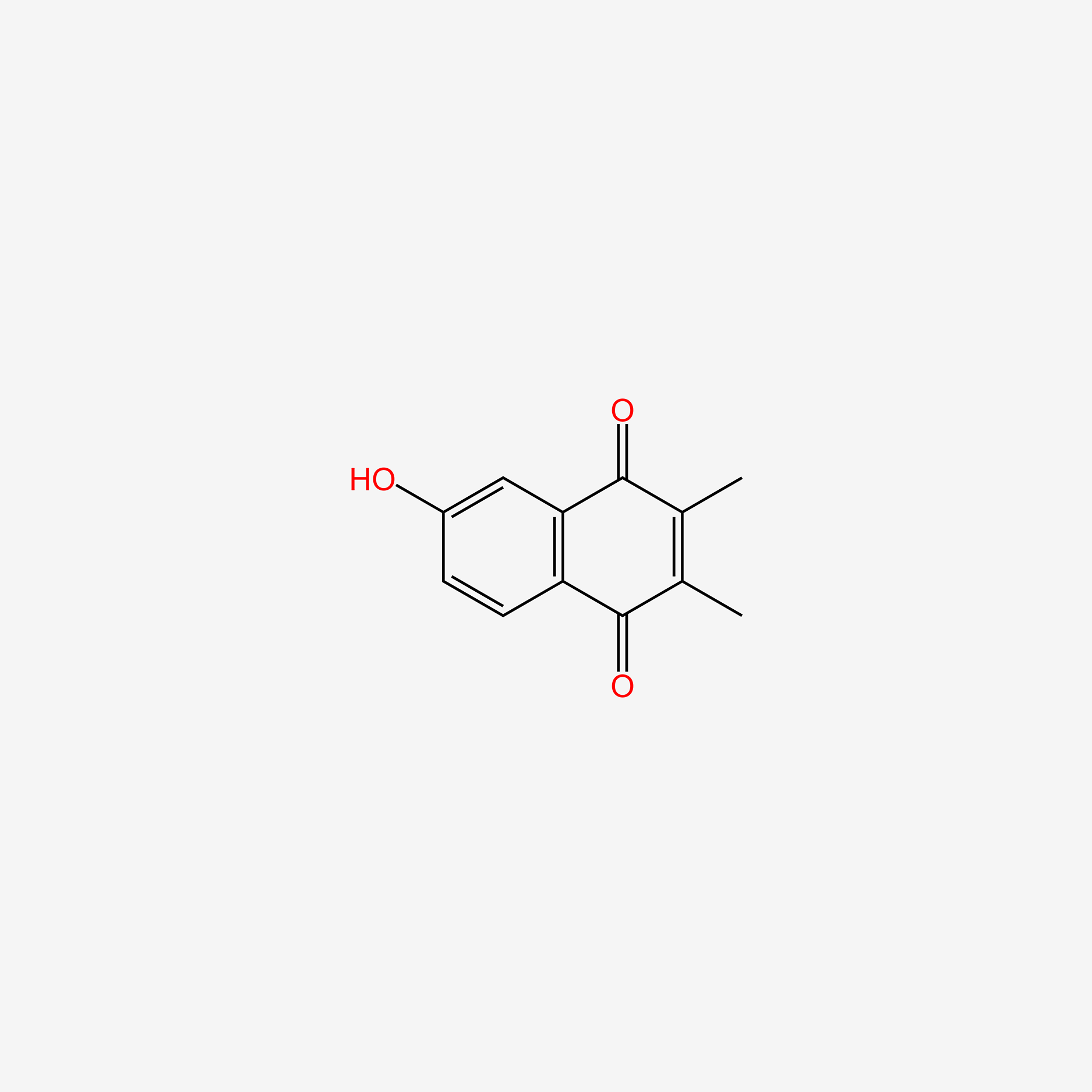 |
0.250 | D0O6KE | 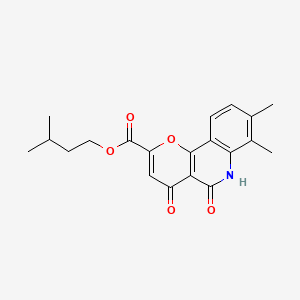 |
0.235 | ||
| ENC005178 | 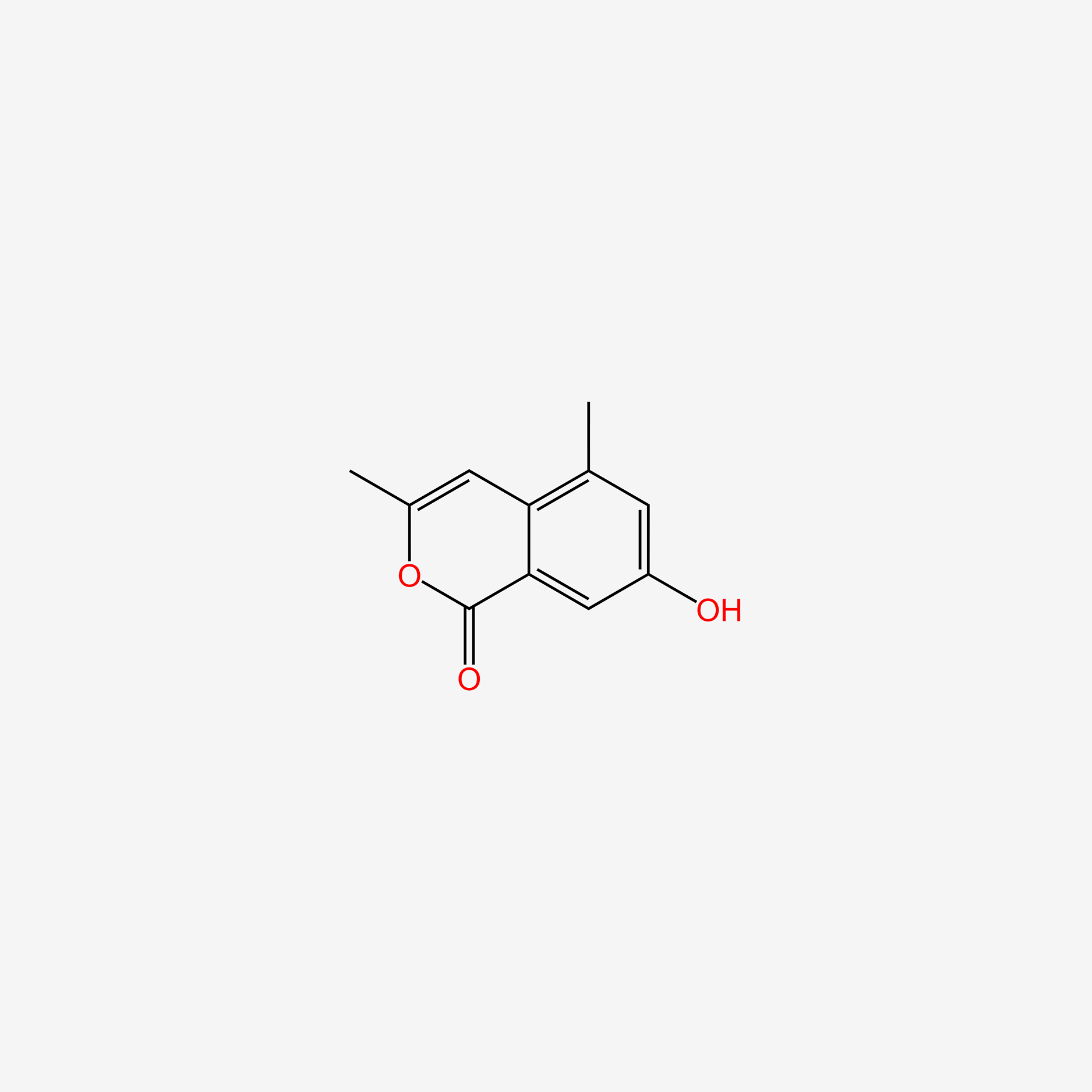 |
0.239 | D05LEO | 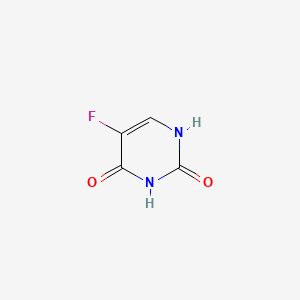 |
0.233 | ||
| ENC001617 | 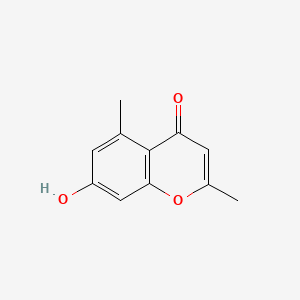 |
0.239 | D05YBZ | 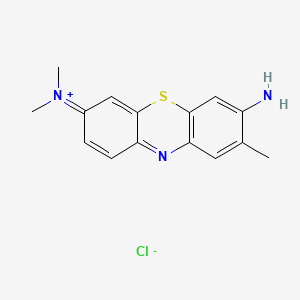 |
0.224 | ||
| ENC001024 | 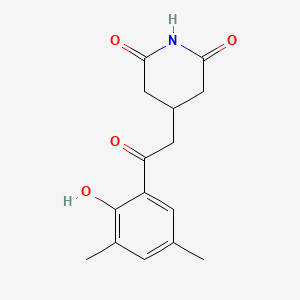 |
0.238 | D0V9YR | 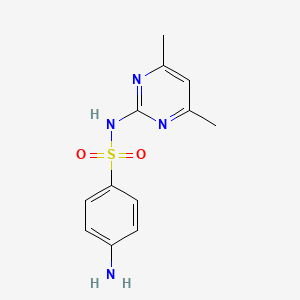 |
0.214 | ||