NPs Basic Information
|
Name |
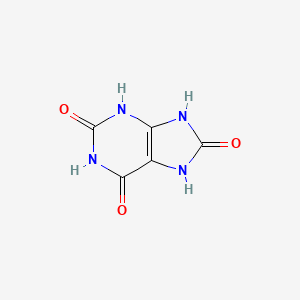
Uric Acid
|
| Molecular Formula | C5H4N4O3 | |
| IUPAC Name* |
7,9-dihydro-3H-purine-2,6,8-trione
|
|
| SMILES |
C12=C(NC(=O)N1)NC(=O)NC2=O
|
|
| InChI |
InChI=1S/C5H4N4O3/c10-3-1-2(7-4(11)6-1)8-5(12)9-3/h(H4,6,7,8,9,10,11,12)
|
|
| InChIKey |
LEHOTFFKMJEONL-UHFFFAOYSA-N
|
|
| Synonyms |
uric acid; 69-93-2; urate; Lithic acid; 2,6,8-trioxypurine; 2,6,8-trihydroxypurine; 8-hydroxyxanthine; 7,9-Dihydro-1H-purine-2,6,8(3H)-trione; 2,6,8-Trioxopurine; 1H-Purine-2,6,8-triol; 1H-Purine-2,6,8(3H)-trione, 7,9-dihydro-; Purine-2,6,8(1H,3H,9H)-trione; 7,9-dihydro-3H-purine-2,6,8-trione; Uricum acidum; AI3-15432; 2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione; NSC 3975; 1H-purine-2,6,8(3H,7H,9H)-trione; NSC-3975; MFCD00005712; Idelalisib metabolite m54; CHEMBL792; 9H-purine-2,6,8-triol; CHEBI:17775; 268B43MJ25; 2,6-dihydroxy-7,9-dihydropurin-8-one; NCGC00181032-01; 8-Hydroxy-3,9-Dihydro-1h-Purine-2,6-Dione; 6,8-Dioxo-6,7,8,9-tetrahydro-1H-purin-2-olate; trioxopurine; URC; EINECS 200-720-7; Lithate; hypoxanthinediol; uric-acid; UNII-268B43MJ25; 2,8-Trioxopurine; 2,8-Trioxypurine; 8HX; Uric acid (8CI); 2,8-Trihydroxypurine; 1l5s; 2,6,8-trihydroxypurin; Purine-2,6,8-triol; 1H-Purine-2,8-triol; Uric acid, 99.0%; URIC ACID [MI]; URIC ACID [INCI]; bmse000126; SCHEMBL7933; 7H-purine-2,6,8-triol; DSSTox_CID_22508; DSSTox_RID_80044; DSSTox_GSID_42508; URICUM ACIDUM [HPUS]; (S)-3-Aminoquinuclidine HCl; GTPL4731; DTXSID3042508; SCHEMBL15777793; SCHEMBL17081907; CHEBI:46811; CHEBI:46814; CHEBI:46817; CHEBI:46823; CHEBI:62589; NSC3975; Uric acid, >=99%, crystalline; HMS3604N17; AMY23430; BCP28980; HY-B2130; Purine-2,8(1H,3H,9H)-trione; ZINC2041003; Tox21_113563; BDBM50325824; s3955; STL185577; AKOS000118731; Purine-3,6,8(1H,3H,9H)-trione; CCG-339700; DB08844; Uric acid, NIST(R) SRM(R) 913b; CAS-69-93-2; purine-2,6,8-(1H,3H,9H)-trione; Uric acid, BioXtra, >=99% (HPLC); AS-56119; SY057305; 6-hydroxy-1H-purine-2,8(7H,9H)-dione; DB-055359; 2,6,8-Trioxypurine;2,6,8-Trihydroxypurine; 2,6-dihydroxy-7,9-dihydro-8H-purin-8-one; CS-0020287; FT-0631301; U0018; 1H-Purine-2,8(3H)-trione, 7,9-dihydro-; EN300-19268; 7,9-Dihydro-3H-purine-2,6,8-trione(Urate); C00366; U-6050; 1H-Purine-2,6,8-triol 2,6,8-Trihydroxypurine; 7,9-Dihydro-3H-purine-2,6,8-trione(uric acid); A866713; Q105522; SR-01000945208; SR-01000945208-1; 565FF3AF-8AFA-4EE9-9FC4-6B119784A5BB; 1H-Purine-2,6,8(3H)-trione, 7,9-dihydro- (9CI); Z104473370
|
|
| CAS | 69-93-2 | |
| PubChem CID | 1175 | |
| ChEMBL ID | CHEMBL792 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 168.11 | ALogp: | -1.9 |
| HBD: | 4 | HBA: | 3 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 99.3 | Aromatic Rings: | 2 |
| Heavy Atoms: | 12 | QED Weighted: | 0.396 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.893 | MDCK Permeability: | 0.00004840 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.251 |
| Human Intestinal Absorption (HIA): | 1 | 20% Bioavailability (F20%): | 0.256 |
| 30% Bioavailability (F30%): | 0.042 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.117 | Plasma Protein Binding (PPB): | 14.34% |
| Volume Distribution (VD): | 0.516 | Fu: | 67.00% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.036 | CYP1A2-substrate: | 0.992 |
| CYP2C19-inhibitor: | 0.074 | CYP2C19-substrate: | 0.039 |
| CYP2C9-inhibitor: | 0.01 | CYP2C9-substrate: | 0.01 |
| CYP2D6-inhibitor: | 0.001 | CYP2D6-substrate: | 0.002 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.425 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.628 | Half-life (T1/2): | 0.971 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.022 | Human Hepatotoxicity (H-HT): | 0.631 |
| Drug-inuced Liver Injury (DILI): | 0.994 | AMES Toxicity: | 0.033 |
| Rat Oral Acute Toxicity: | 0.981 | Maximum Recommended Daily Dose: | 0.007 |
| Skin Sensitization: | 0.174 | Carcinogencity: | 0.383 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.843 |
| Respiratory Toxicity: | 0.979 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000063 | 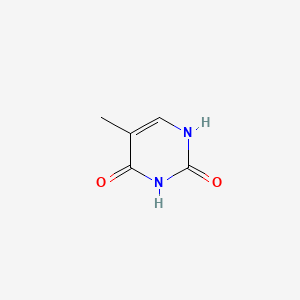 |
0.295 | D05LEO | 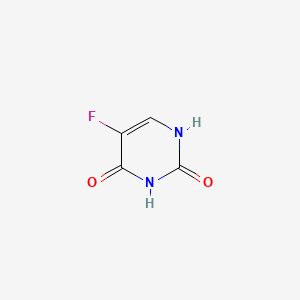 |
0.295 | ||
| ENC001638 | 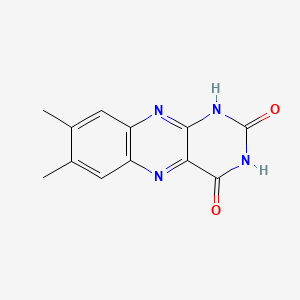 |
0.258 | D09AMZ | 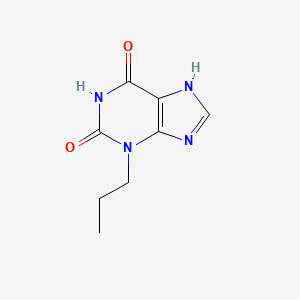 |
0.241 | ||
| ENC000065 | 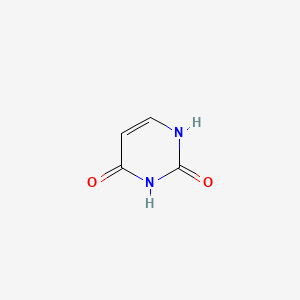 |
0.222 | D0WB9V | 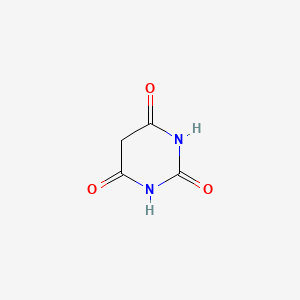 |
0.239 | ||
| ENC000997 | 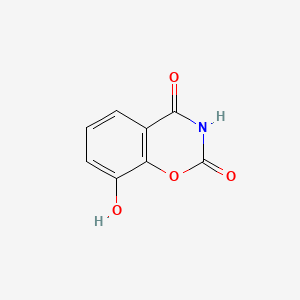 |
0.211 | D0Y8PT | 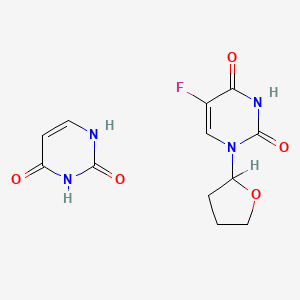 |
0.237 | ||
| ENC003436 | 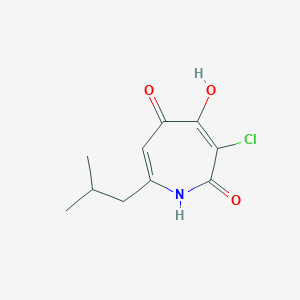 |
0.200 | D0J9UN | 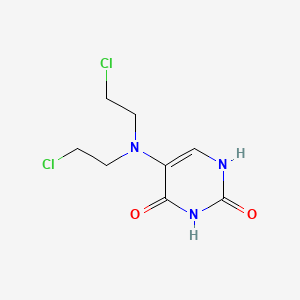 |
0.213 | ||
| ENC002826 | 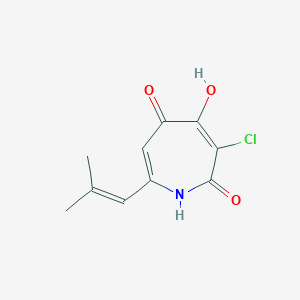 |
0.200 | D0E0SW | 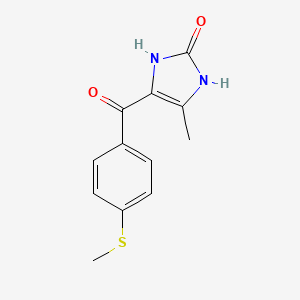 |
0.212 | ||
| ENC002824 | 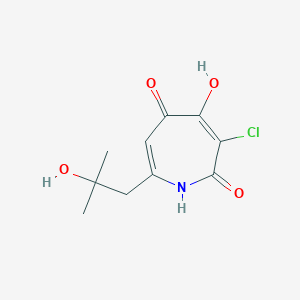 |
0.194 | D0E0WQ | 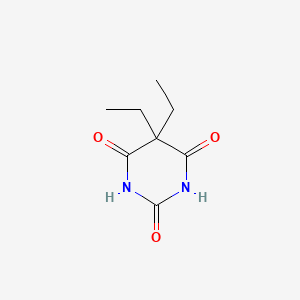 |
0.196 | ||
| ENC002446 | 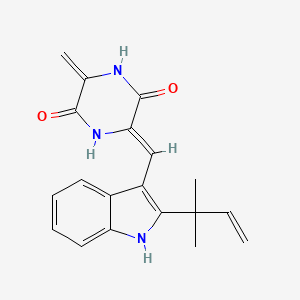 |
0.193 | D0I0DS | 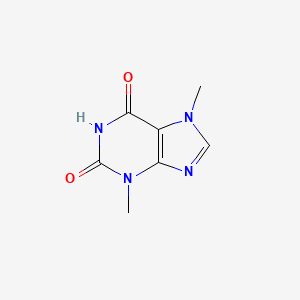 |
0.193 | ||
| ENC004721 | 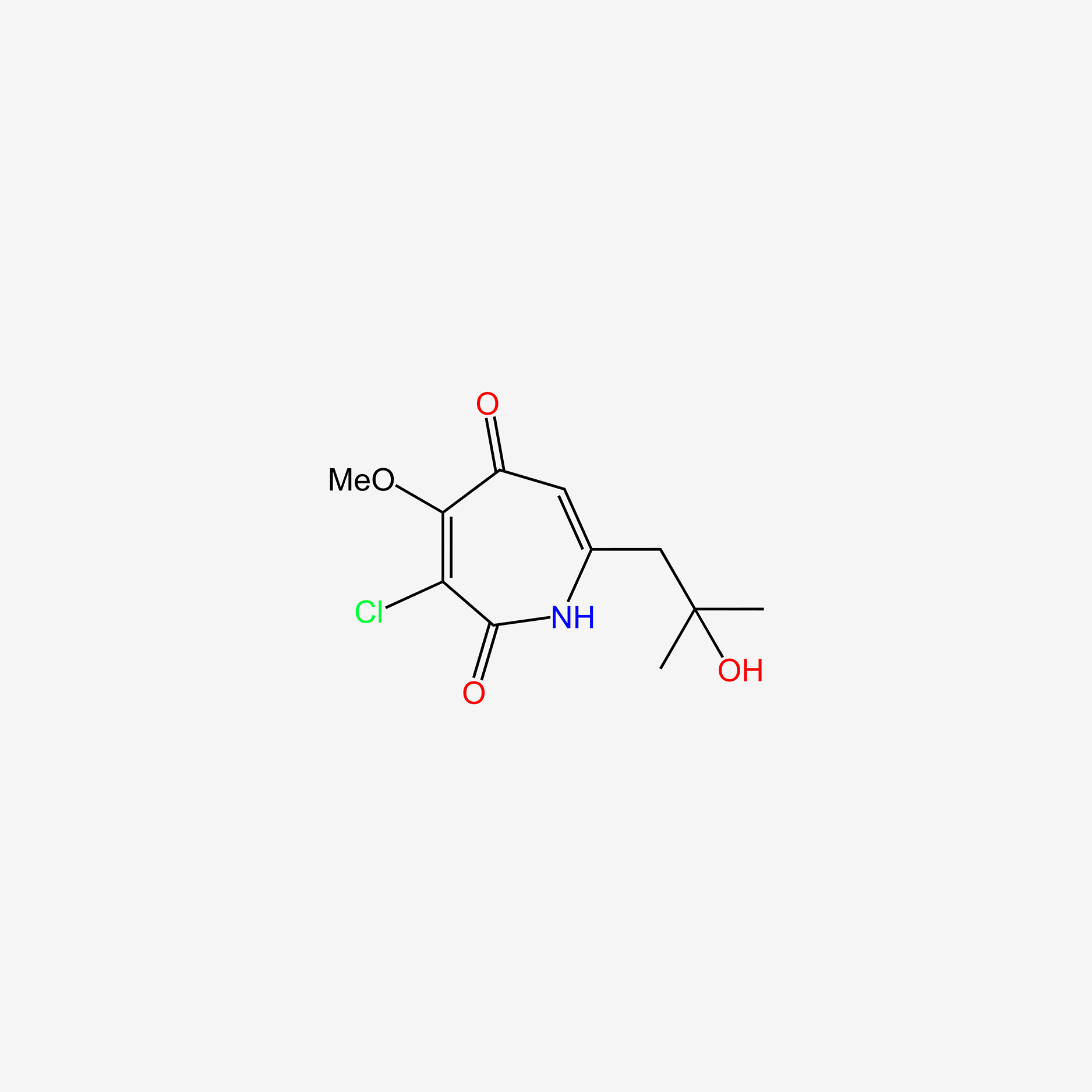 |
0.185 | D0A4JK | 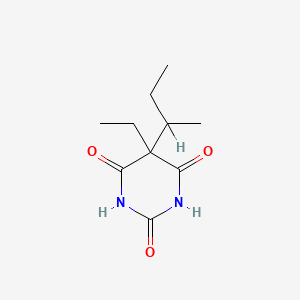 |
0.180 | ||
| ENC004720 | 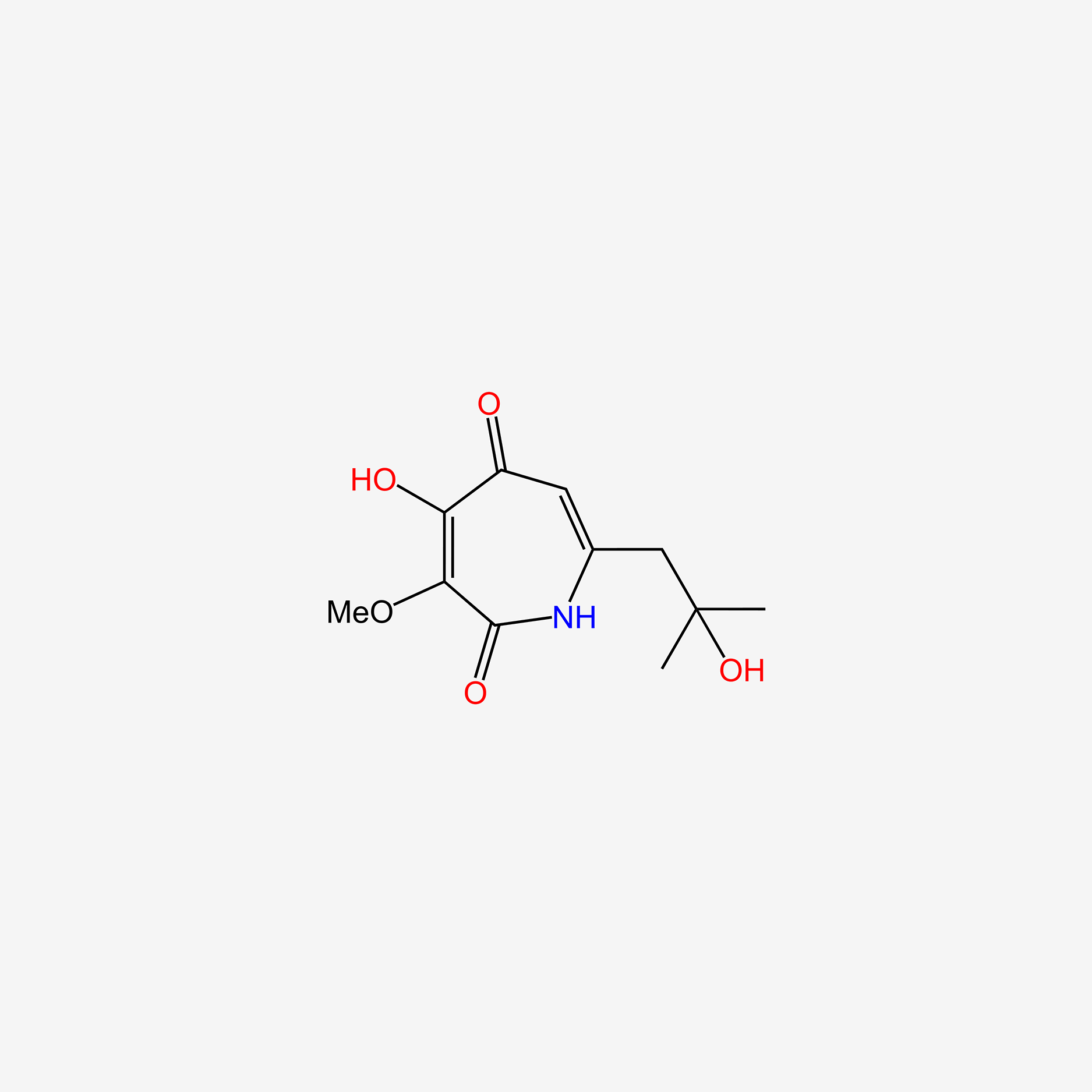 |
0.185 | D05TMQ |  |
0.180 | ||