NPs Basic Information
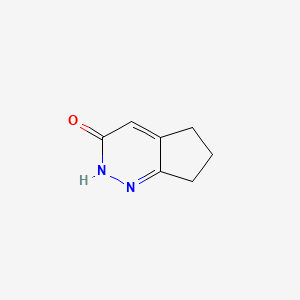
|
Name |
2,5,6,7-tetrahydro-3H-cyclopenta[c]pyridazin-3-one
|
| Molecular Formula | C7H8N2O | |
| IUPAC Name* |
2,5,6,7-tetrahydrocyclopenta[c]pyridazin-3-one
|
|
| SMILES |
C1CC2=CC(=O)NN=C2C1
|
|
| InChI |
InChI=1S/C7H8N2O/c10-7-4-5-2-1-3-6(5)8-9-7/h4H,1-3H2,(H,9,10)
|
|
| InChIKey |
HZLVYABRXDPPSA-UHFFFAOYSA-N
|
|
| Synonyms |
122001-78-9; 2,5,6,7-tetrahydro-3H-cyclopenta[c]pyridazin-3-one; 6,7-Dihydro-2H-cyclopenta[c]pyridazin-3(5H)-one; 6,7-Dihydro-5H-cyclopenta[c]pyridazin-3-ol; 2H,3H,5H,6H,7H-cyclopenta[c]pyridazin-3-one; 2,5,6,7-tetrahydrocyclopenta[c]pyridazin-3-one; 2,5,6,7-TETRAHYDRO-CYCLOPENTA[C]PYRIDAZIN-3-ONE; MFCD07757617; 3H-Cyclopenta[c]pyridazin-3-one, 2,5,6,7-tetrahydro-; SCHEMBL20092675; DTXSID40409432; ALBB-028380; ZINC4342676; STK785697; AKOS005621180; AKOS023410365; AS-30198; SY180609; CS-0075473; EN300-60243; F1967-2558; F8888-0981; Z935132918; 2,5,6,7-tetrahydro-3H-cyclopenta[c]pyridazin-3-one(SALTDATA: FREE)
|
|
| CAS | 122001-78-9 | |
| PubChem CID | 5200288 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.15 | ALogp: | -0.1 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 41.5 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.569 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.654 | MDCK Permeability: | 0.00001580 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.449 |
| Human Intestinal Absorption (HIA): | 0.011 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.386 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.977 | Plasma Protein Binding (PPB): | 24.58% |
| Volume Distribution (VD): | 0.953 | Fu: | 62.50% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.101 | CYP1A2-substrate: | 0.831 |
| CYP2C19-inhibitor: | 0.081 | CYP2C19-substrate: | 0.059 |
| CYP2C9-inhibitor: | 0.029 | CYP2C9-substrate: | 0.907 |
| CYP2D6-inhibitor: | 0.05 | CYP2D6-substrate: | 0.765 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.112 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.599 | Half-life (T1/2): | 0.649 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.023 | Human Hepatotoxicity (H-HT): | 0.245 |
| Drug-inuced Liver Injury (DILI): | 0.654 | AMES Toxicity: | 0.118 |
| Rat Oral Acute Toxicity: | 0.125 | Maximum Recommended Daily Dose: | 0.595 |
| Skin Sensitization: | 0.145 | Carcinogencity: | 0.261 |
| Eye Corrosion: | 0.004 | Eye Irritation: | 0.617 |
| Respiratory Toxicity: | 0.437 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000345 | 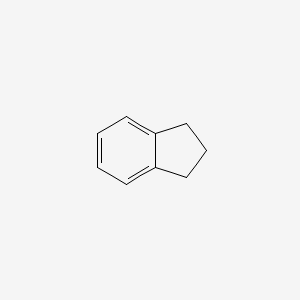 |
0.273 | D0E6YQ | 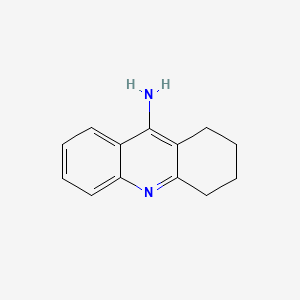 |
0.217 | ||
| ENC000518 | 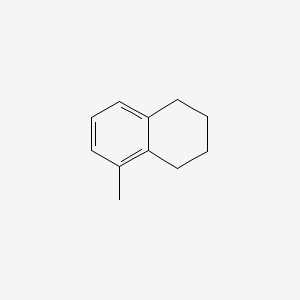 |
0.245 | D05IHU | 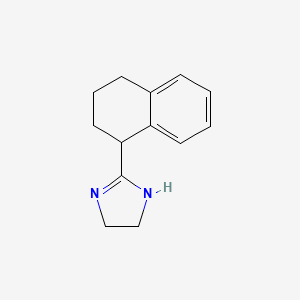 |
0.213 | ||
| ENC000393 | 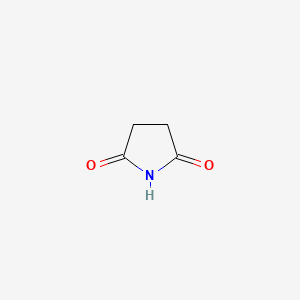 |
0.231 | D00EEL | 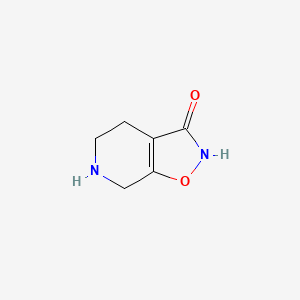 |
0.208 | ||
| ENC003872 | 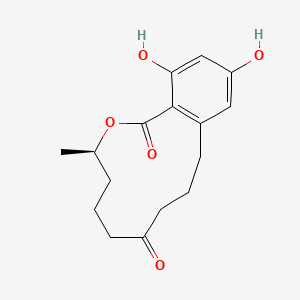 |
0.225 | D01JMC | 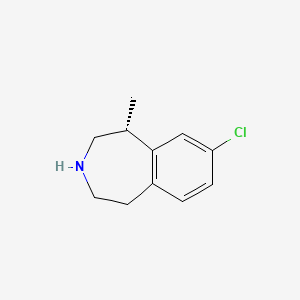 |
0.200 | ||
| ENC000450 | 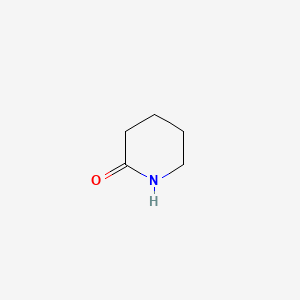 |
0.225 | D00ETS |  |
0.190 | ||
| ENC000038 | 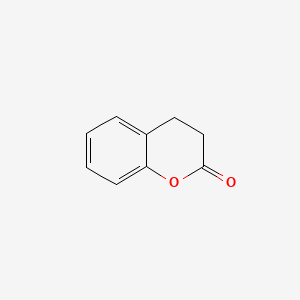 |
0.220 | D0V1UW | 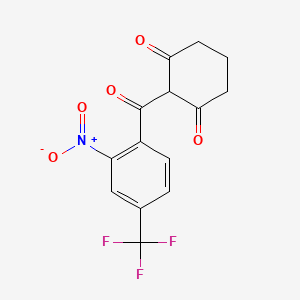 |
0.184 | ||
| ENC000476 | 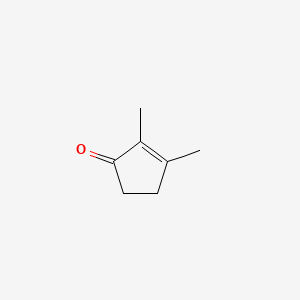 |
0.220 | D04KYY | 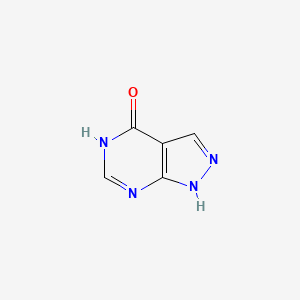 |
0.184 | ||
| ENC000753 | 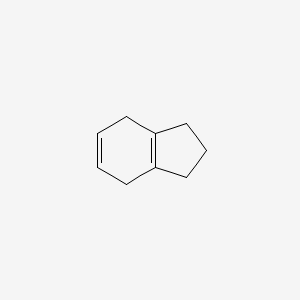 |
0.217 | D07GRH | 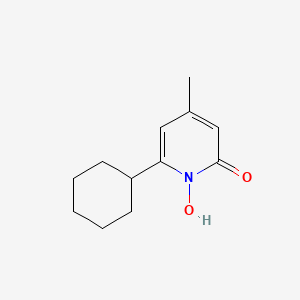 |
0.183 | ||
| ENC003715 |  |
0.216 | D0N8DP |  |
0.182 | ||
| ENC003479 |  |
0.212 | D00MIN |  |
0.180 | ||