NPs Basic Information
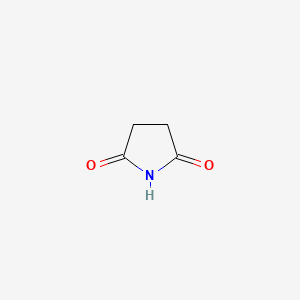
|
Name |
Succinimide
|
| Molecular Formula | C4H5NO2 | |
| IUPAC Name* |
pyrrolidine-2,5-dione
|
|
| SMILES |
C1CC(=O)NC1=O
|
|
| InChI |
InChI=1S/C4H5NO2/c6-3-1-2-4(7)5-3/h1-2H2,(H,5,6,7)
|
|
| InChIKey |
KZNICNPSHKQLFF-UHFFFAOYSA-N
|
|
| Synonyms |
SUCCINIMIDE; 123-56-8; pyrrolidine-2,5-dione; 2,5-Pyrrolidinedione; Butanimide; Succinic acid imide; 2,5-Dioxopyrrolidine; 2,5-Diketopyrrolidine; Succinic imide; Succinimide-sauba; 3,4-Dihydropyrrole-2,5-dione; Dihydro-3-pyrroline-2,5-dione; 3,4-Dihydropyrrolidine; NSC 11204; SUCCIMIDE; Mercuric imidosuccinate; CHEBI:9307; NSC-11204; Mercury, disuccinimido-; WLN: T5VMVTJ; 2, mercury(2+) salt; 10X90O3503; 584-43-0; Succinimide, mercury(2+) salt; MERCURIC SUCCINIMIDE; 25950-42-9; NSC-38417; NSC-41221; EINECS 204-635-6; MFCD00005495; BRN 0108440; succinimid; cis-succinimide; AI3-08539; UNII-10X90O3503; Succinimide (SI); 2,5dioxopyrrolidine; Lubrizol 6406; pyrroldine-2,5-dione; succinimide(butanimide); SUCCINIMIDE [MI]; Succinimide pharbiol (TN); EC 204-635-6; DSSTox_CID_30181; SUCCINIMIDE [MART.]; DSSTox_GSID_51629; SUCCINIMIDE [WHO-DD]; 5-21-09-00438 (Beilstein Handbook Reference); BDBM7814; CHEMBL275661; DTXSID8051629; NSC11204; NSC13114; NSC38417; NSC41221; NSC49152; ZINC5133396; Tox21_303851; NSC-13114; NSC-49152; STL163344; AKOS000118776; DB13376; NCGC00357119-01; BP-21153; CAS-123-56-8; BB 0322715; FT-0658724; Succinimide, Vetec(TM) reagent grade, 98%; EN300-17963; C07273; D08532; Q419430; Z57127349; F1908-0169; 1BBF5533-77F1-4B20-96F3-0BB022C36CD3
|
|
| CAS | 123-56-8 | |
| PubChem CID | 11439 | |
| ChEMBL ID | CHEMBL275661 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 99.09 | ALogp: | -1.0 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 46.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 7 | QED Weighted: | 0.43 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.096 | MDCK Permeability: | 0.00000838 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.014 | 20% Bioavailability (F20%): | 0.937 |
| 30% Bioavailability (F30%): | 0.502 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.066 | Plasma Protein Binding (PPB): | 27.14% |
| Volume Distribution (VD): | 0.976 | Fu: | 70.60% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.017 | CYP1A2-substrate: | 0.188 |
| CYP2C19-inhibitor: | 0.034 | CYP2C19-substrate: | 0.054 |
| CYP2C9-inhibitor: | 0.012 | CYP2C9-substrate: | 0.921 |
| CYP2D6-inhibitor: | 0.01 | CYP2D6-substrate: | 0.719 |
| CYP3A4-inhibitor: | 0.01 | CYP3A4-substrate: | 0.081 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.409 | Half-life (T1/2): | 0.881 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.009 | Human Hepatotoxicity (H-HT): | 0.193 |
| Drug-inuced Liver Injury (DILI): | 0.492 | AMES Toxicity: | 0.186 |
| Rat Oral Acute Toxicity: | 0.658 | Maximum Recommended Daily Dose: | 0.007 |
| Skin Sensitization: | 0.094 | Carcinogencity: | 0.703 |
| Eye Corrosion: | 0.341 | Eye Irritation: | 0.654 |
| Respiratory Toxicity: | 0.717 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001020 | 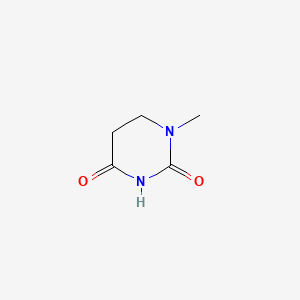 |
0.433 | D0K8IX | 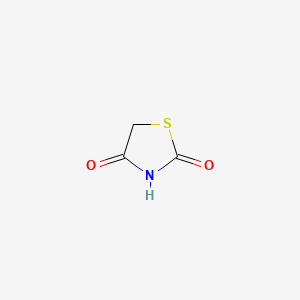 |
0.357 | ||
| ENC005486 | 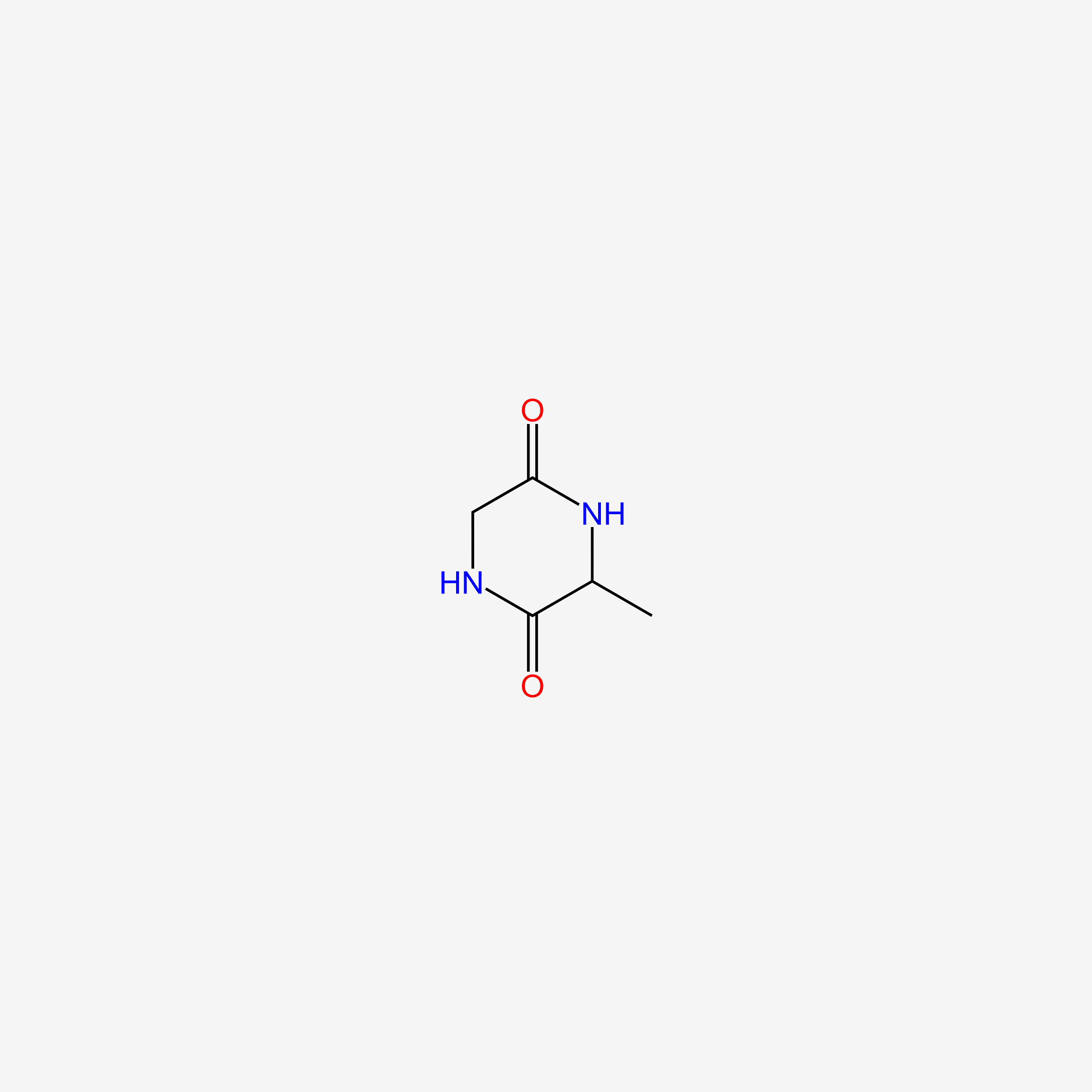 |
0.265 | D0WB9V | 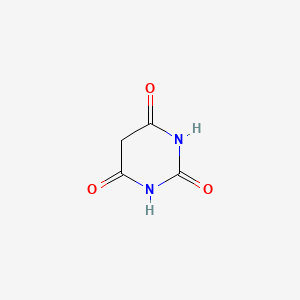 |
0.344 | ||
| ENC000450 | 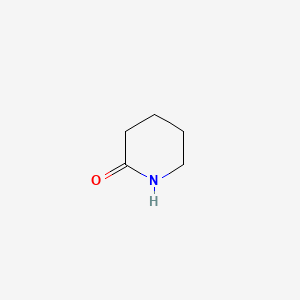 |
0.258 | D0Q4XQ | 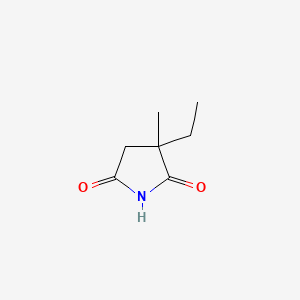 |
0.286 | ||
| ENC000476 | 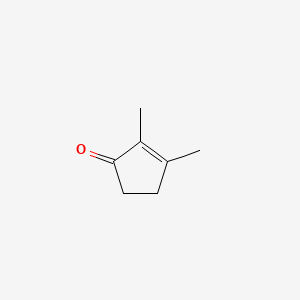 |
0.250 | D0Z9NZ | 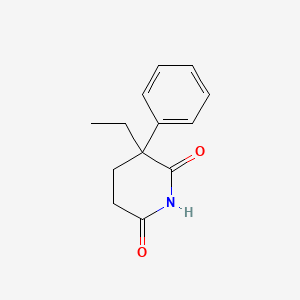 |
0.255 | ||
| ENC001229 | 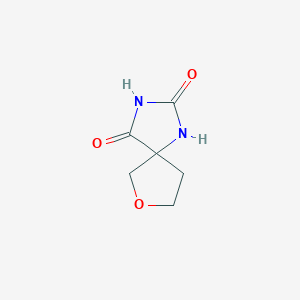 |
0.250 | D0M6DO | 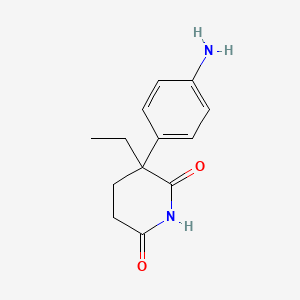 |
0.245 | ||
| ENC003479 | 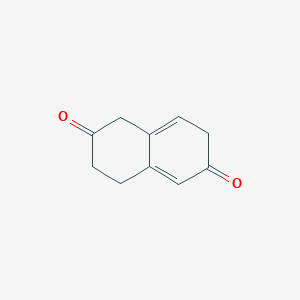 |
0.233 | D0Z8AA | 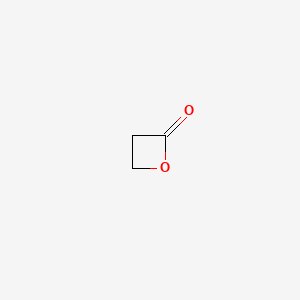 |
0.240 | ||
| ENC001520 | 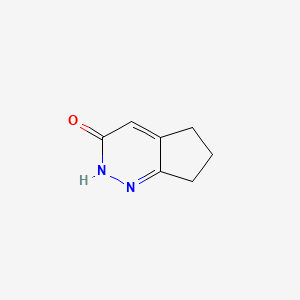 |
0.231 | D07XVN | 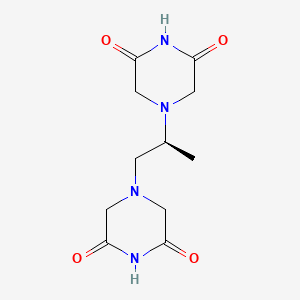 |
0.224 | ||
| ENC002343 | 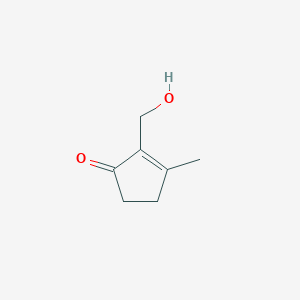 |
0.229 | D0Q5NX | 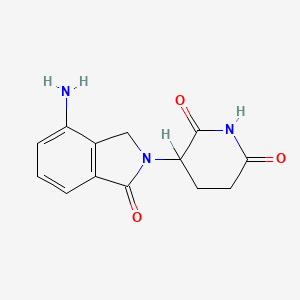 |
0.220 | ||
| ENC006064 | 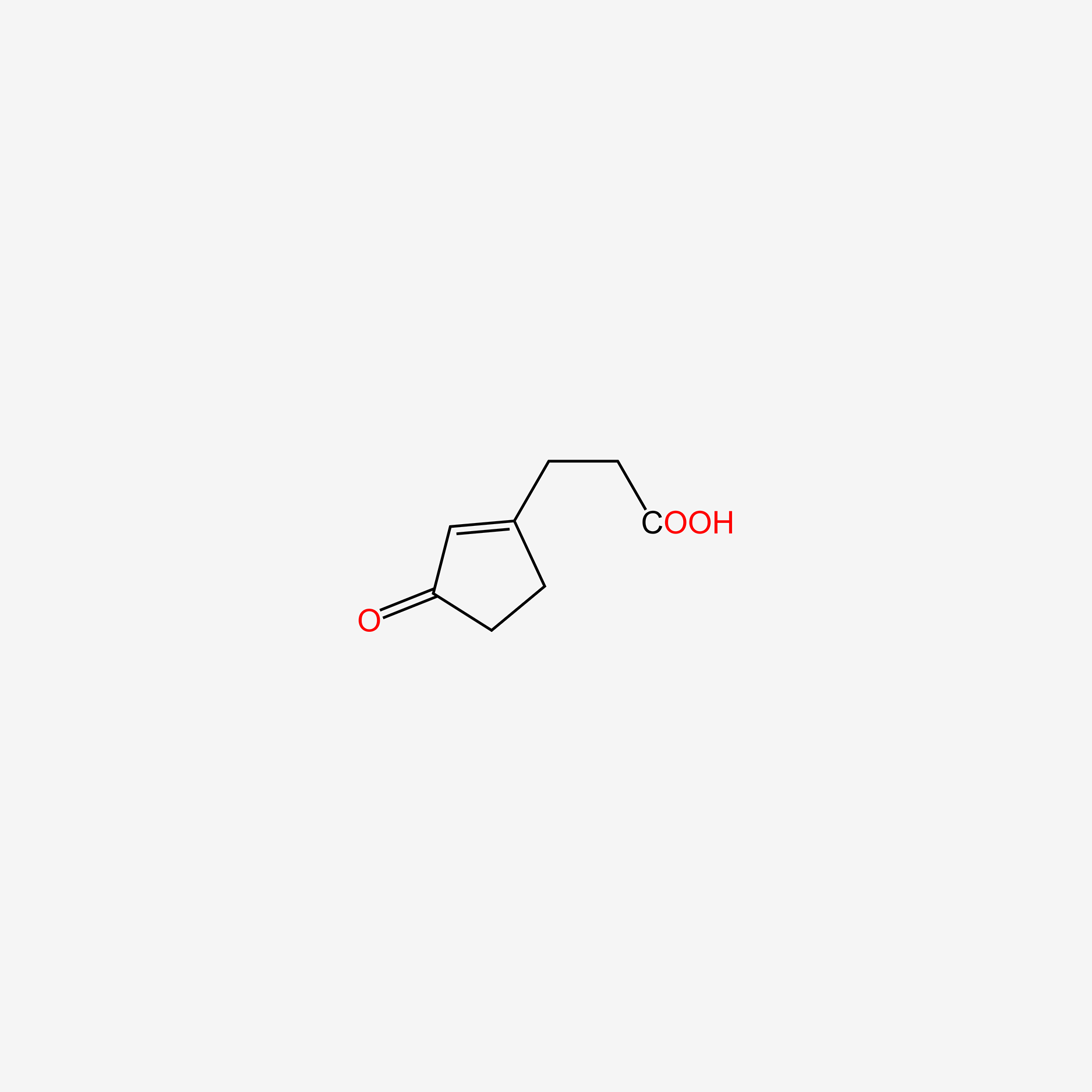 |
0.225 | D0U7GK | 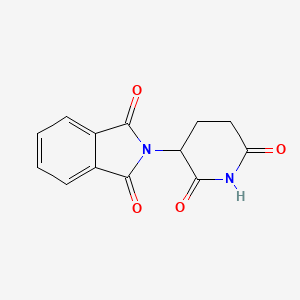 |
0.220 | ||
| ENC005741 | 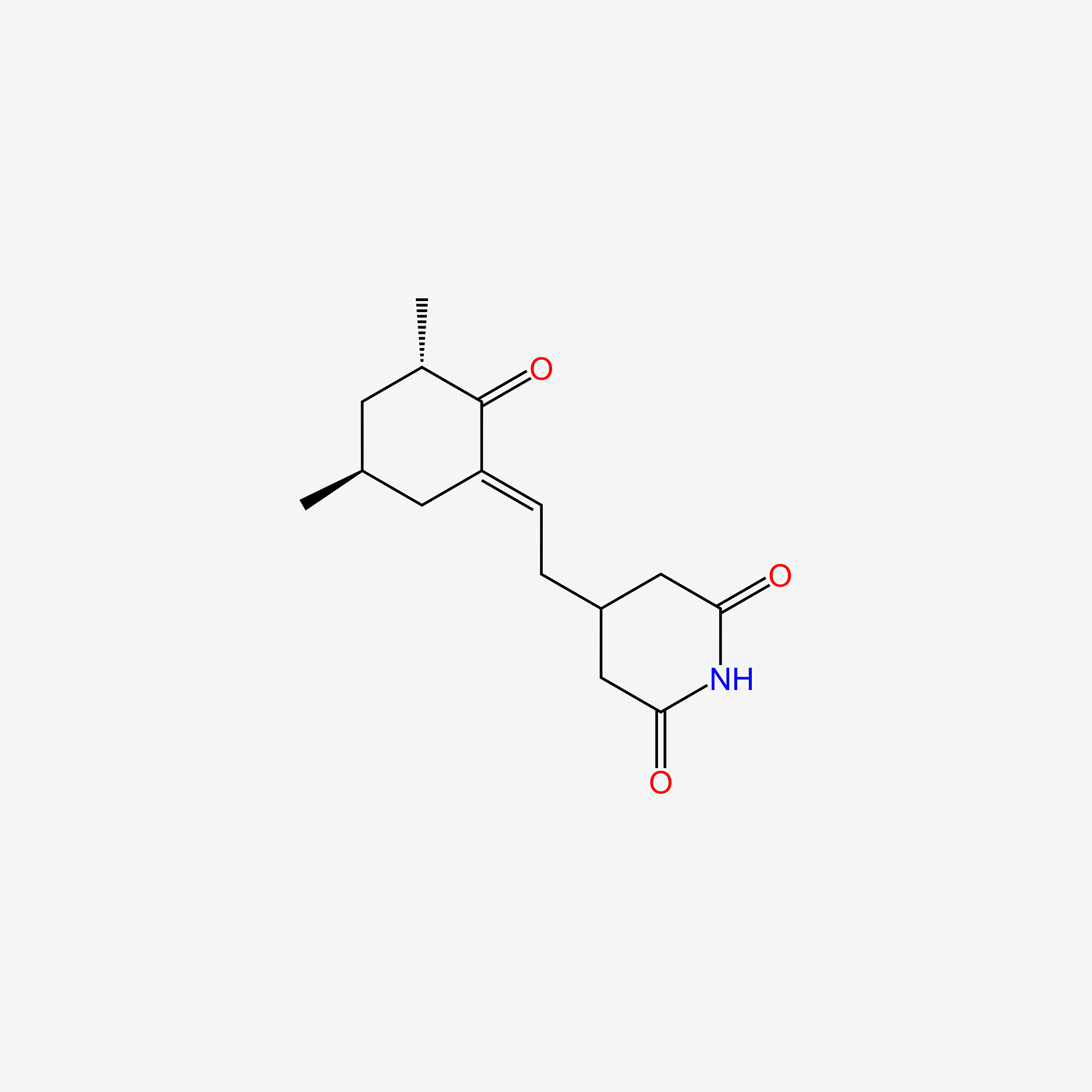 |
0.224 | D0A3ZU | 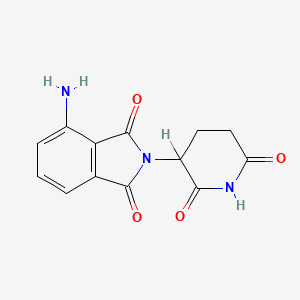 |
0.213 | ||