NPs Basic Information
|
Name |
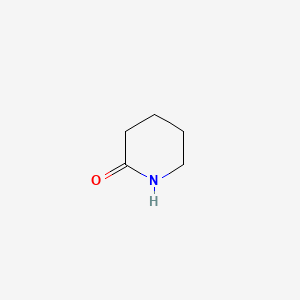
2-Piperidone
|
| Molecular Formula | C5H9NO | |
| IUPAC Name* |
piperidin-2-one
|
|
| SMILES |
C1CCNC(=O)C1
|
|
| InChI |
InChI=1S/C5H9NO/c7-5-3-1-2-4-6-5/h1-4H2,(H,6,7)
|
|
| InChIKey |
XUWHAWMETYGRKB-UHFFFAOYSA-N
|
|
| Synonyms |
2-Piperidone; piperidin-2-one; 675-20-7; DELTA-VALEROLACTAM; 2-Piperidinone; Valerolactim; 5-Pentanolactam; Piperidone; Piperidinone; Piperidon; Valerolactam; alpha-Piperidone; Piperidone-2; PIPERIDONE,2-; .alpha.-Piperidone; 2-oxopiperidine; 2-Azacyclohexanone; Piperidon [German]; Pentanoic acid, 5-amino-, lactam; .delta.-Valerolactam; A-Piperidone; 2-oxo-piperidine; NSC 2305; NSC 18894; WLN0GQQ6EK; MFCD00006037; CHEMBL12193; CHEBI:77761; NSC-2305; NSC18894; Piperidon (german); NSC-18894; Pentanoic acid, lactam; WLN: T6NVTJ; 25036-00-4; Piperidones; Piperidone-2 [French]; d-Valerolactam; 5-pentanelactam; UNII-WLN0GQQ6EK; EINECS 211-622-9; ketopiperidine; oxopiperidine; 2-piperadinone; AI3-33342; d-Valero-lactam; piperadine-2-one; piperidine-2-one; delta -Valerolactam; V1L; 27154-43-4; 5-amino-lactam-Pentanoate; delta-Valerolactam, 98%; BDBM10; 5-amino-lactam-Pentanoic acid; piperidin-2-one;2-Piperidone; DTXSID1060976; NSC2305; BCP00878; ZINC3860817; BBL027557; STL281850; AKOS005206867; CS-W022933; HY-W042193; SB41073; AC-15619; AC-33837; NCI60_001574; SY011119; DB-031244; A9047; AM20100626; FT-0613368; P0455; EN300-24025; F10312; 675V207; Q4596918; W-104710; F0001-1780; Z168817684
|
|
| CAS | 675-20-7 | |
| PubChem CID | 12665 | |
| ChEMBL ID | CHEMBL12193 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 99.13 | ALogp: | -0.5 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 29.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 7 | QED Weighted: | 0.475 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.355 | MDCK Permeability: | 0.00003590 |
| Pgp-inhibitor: | 0.005 | Pgp-substrate: | 0.041 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.052 |
| 30% Bioavailability (F30%): | 0.4 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.996 | Plasma Protein Binding (PPB): | 7.52% |
| Volume Distribution (VD): | 1.139 | Fu: | 83.57% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.174 | CYP1A2-substrate: | 0.249 |
| CYP2C19-inhibitor: | 0.062 | CYP2C19-substrate: | 0.542 |
| CYP2C9-inhibitor: | 0.019 | CYP2C9-substrate: | 0.312 |
| CYP2D6-inhibitor: | 0.016 | CYP2D6-substrate: | 0.353 |
| CYP3A4-inhibitor: | 0.014 | CYP3A4-substrate: | 0.209 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.962 | Half-life (T1/2): | 0.808 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.015 | Human Hepatotoxicity (H-HT): | 0.07 |
| Drug-inuced Liver Injury (DILI): | 0.044 | AMES Toxicity: | 0.025 |
| Rat Oral Acute Toxicity: | 0.239 | Maximum Recommended Daily Dose: | 0.035 |
| Skin Sensitization: | 0.383 | Carcinogencity: | 0.034 |
| Eye Corrosion: | 0.031 | Eye Irritation: | 0.814 |
| Respiratory Toxicity: | 0.058 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001341 | 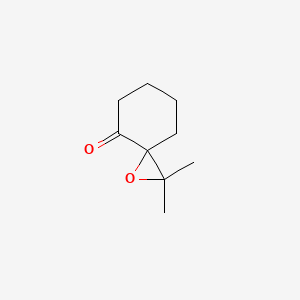 |
0.289 | D0Z8AA | 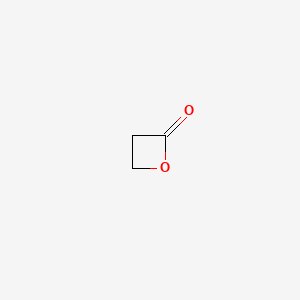 |
0.231 | ||
| ENC000184 | 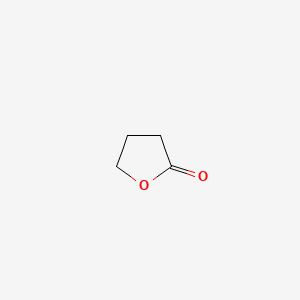 |
0.276 | D00EEL | 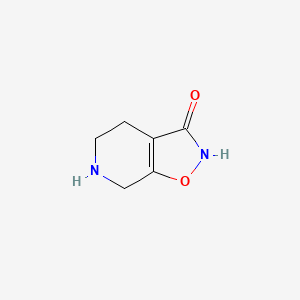 |
0.225 | ||
| ENC001349 | 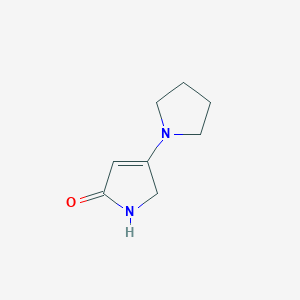 |
0.268 | D00ETS |  |
0.222 | ||
| ENC001712 | 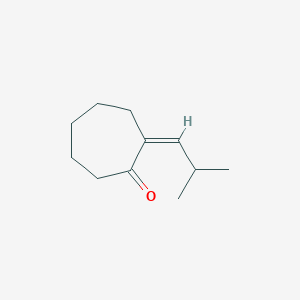 |
0.262 | D03KEK | 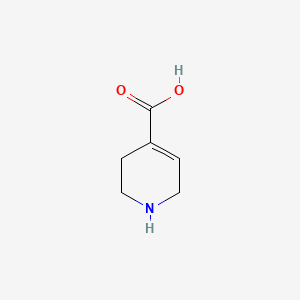 |
0.216 | ||
| ENC000393 | 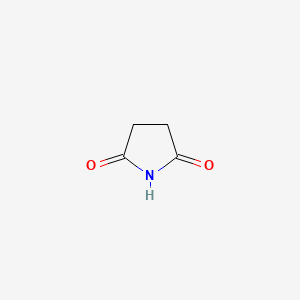 |
0.258 | D03WAJ | 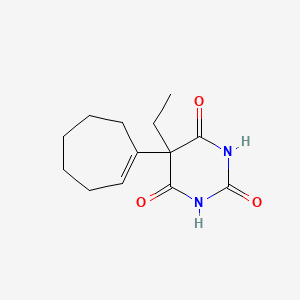 |
0.207 | ||
| ENC001201 | 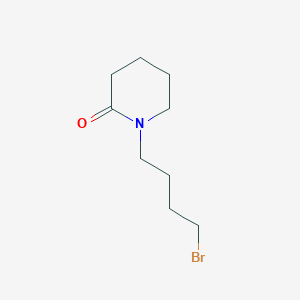 |
0.256 | D0Q4YK | 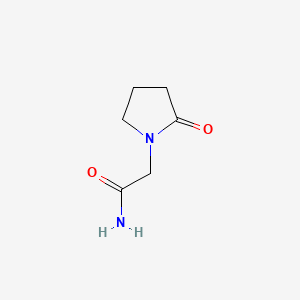 |
0.205 | ||
| ENC000901 | 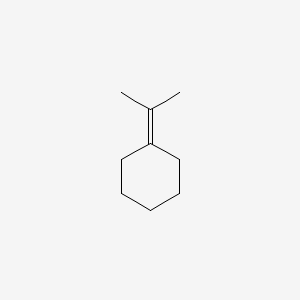 |
0.250 | D0UM7O | 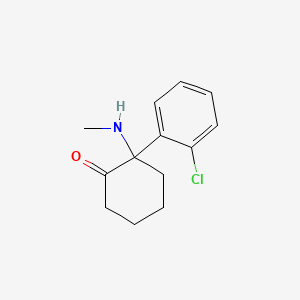 |
0.204 | ||
| ENC000882 | 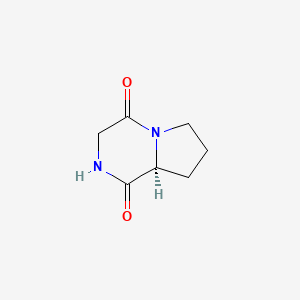 |
0.244 | D0DZ3X | 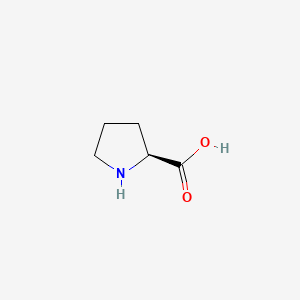 |
0.200 | ||
| ENC000991 | 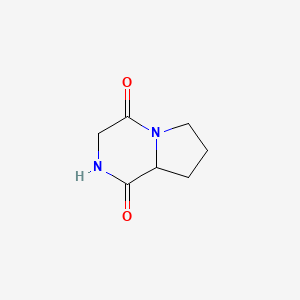 |
0.244 | D02PPN | 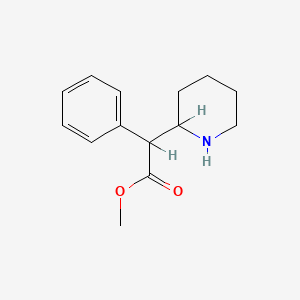 |
0.190 | ||
| ENC001302 | 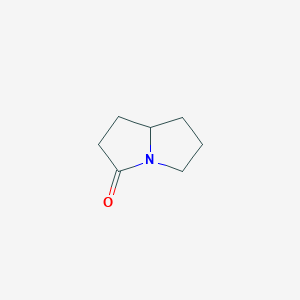 |
0.243 | D0N8DP | 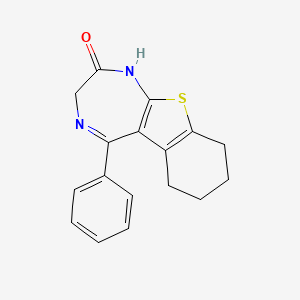 |
0.188 | ||