NPs Basic Information
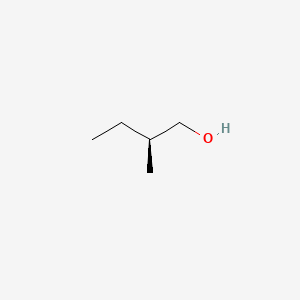
|
Name |
(S)-(-)-2-Methyl-1-butanol
|
| Molecular Formula | C5H12O | |
| IUPAC Name* |
(2S)-2-methylbutan-1-ol
|
|
| SMILES |
CC[C@H](C)CO
|
|
| InChI |
InChI=1S/C5H12O/c1-3-5(2)4-6/h5-6H,3-4H2,1-2H3/t5-/m0/s1
|
|
| InChIKey |
QPRQEDXDYOZYLA-YFKPBYRVSA-N
|
|
| Synonyms |
1565-80-6; (S)-(-)-2-Methyl-1-butanol; (2S)-2-methylbutan-1-ol; (S)-2-Methyl-1-butanol; (S)-2-Methylbutan-1-ol; 1-Butanol, 2-methyl-, (2S)-; 1-Butanol, 2-methyl-, (S)-; (S)-(-)-2-Methylbutanol; 2-Methyl-1-butanol, (-)-; (-)-2-methyl-1-butanol; (2S)-2-methyl-1-butanol; (S)-2-methylbutanol; II2QJB35IC; L-2-Methyl-1-butanol; (-)-2-methylbutanol; active-Amyl Alcohol; UNII-II2QJB35IC; (2S)-2-Methyl-Butan-1-Ol; EINECS 216-366-1; (s)-2-methylbutylalcohol; (S)-2-methylbutyl alcohol; (s)-2-methyl-butan-1-ol; 2-Methyl-1-butanol, (S)-; CHEBI:50625; (-)-(s)-2-methyl-1-butanol; DTXSID20883695; (S)-(-)-2-methylbutan-1-ol; ZINC2040993; MFCD00064299; (S)-(-)-2-Methylbutanol, 99%; D-(-)-2-METHYL-1-BUTANOL; AKOS024256629; CS-0139702; M0170; 2-METHYL-1-BUTANOL (-)-FORM [MI]; EN300-203697; W-108011; Q27122155; (S)-(-)-2-Methylbutanol, >=95.0% (sum of enantiomers, GC)
|
|
| CAS | 1565-80-6 | |
| PubChem CID | 2723602 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 88.15 | ALogp: | 1.2 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 6 | QED Weighted: | 0.541 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.219 | MDCK Permeability: | 0.00007250 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.037 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.031 |
| 30% Bioavailability (F30%): | 0.008 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.956 | Plasma Protein Binding (PPB): | 17.62% |
| Volume Distribution (VD): | 1.027 | Fu: | 79.71% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.376 | CYP1A2-substrate: | 0.678 |
| CYP2C19-inhibitor: | 0.038 | CYP2C19-substrate: | 0.592 |
| CYP2C9-inhibitor: | 0.016 | CYP2C9-substrate: | 0.184 |
| CYP2D6-inhibitor: | 0.016 | CYP2D6-substrate: | 0.234 |
| CYP3A4-inhibitor: | 0.007 | CYP3A4-substrate: | 0.198 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.731 | Half-life (T1/2): | 0.81 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.02 | Human Hepatotoxicity (H-HT): | 0.076 |
| Drug-inuced Liver Injury (DILI): | 0.08 | AMES Toxicity: | 0.014 |
| Rat Oral Acute Toxicity: | 0.32 | Maximum Recommended Daily Dose: | 0.022 |
| Skin Sensitization: | 0.415 | Carcinogencity: | 0.57 |
| Eye Corrosion: | 0.969 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.11 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000307 | 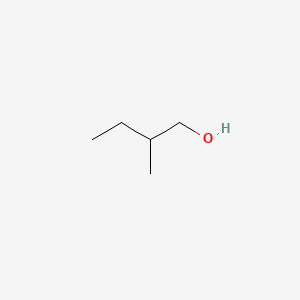 |
1.000 | D00AMQ | 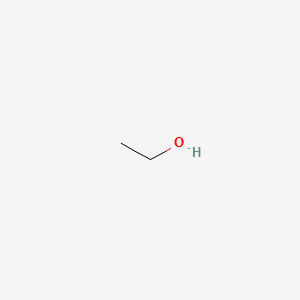 |
0.313 | ||
| ENC000396 | 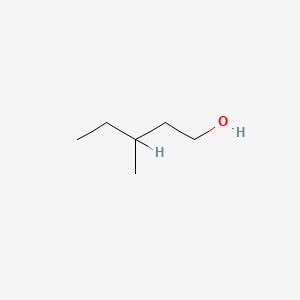 |
0.571 | D0X2IE | 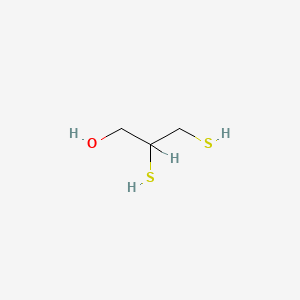 |
0.250 | ||
| ENC000182 |  |
0.500 | D0ZK8H | 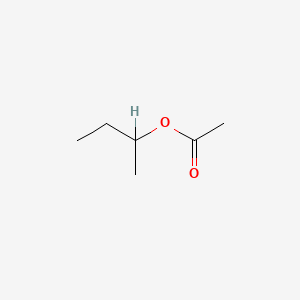 |
0.250 | ||
| ENC000147 | 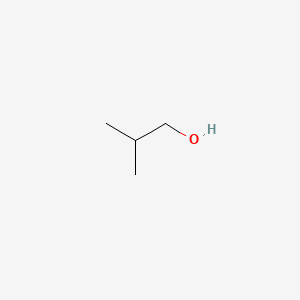 |
0.444 | D08QME | 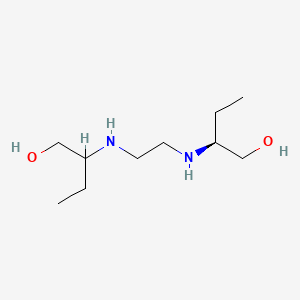 |
0.233 | ||
| ENC000225 | 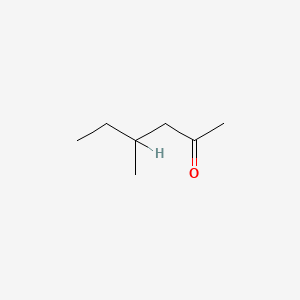 |
0.400 | D0C1QZ | 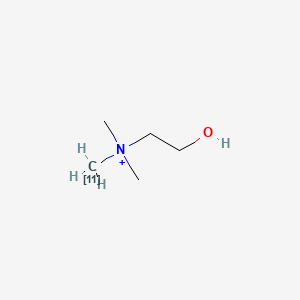 |
0.231 | ||
| ENC001246 | 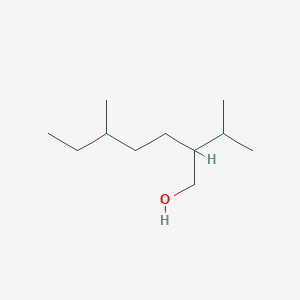 |
0.394 | D0Y3KG | 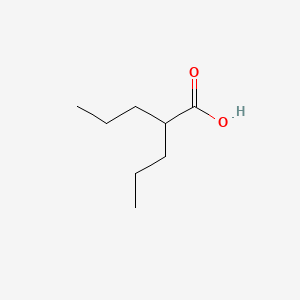 |
0.206 | ||
| ENC000057 | 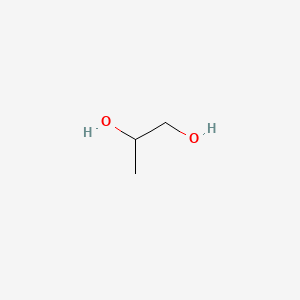 |
0.368 | D00WUF | 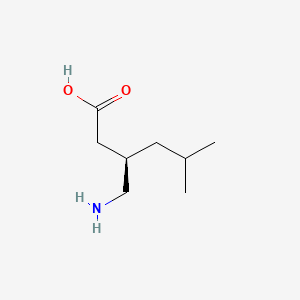 |
0.194 | ||
| ENC001788 | 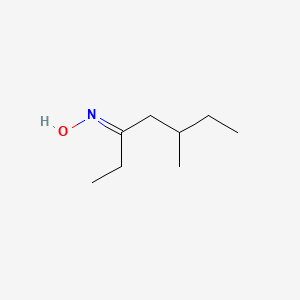 |
0.367 | D02UDJ | 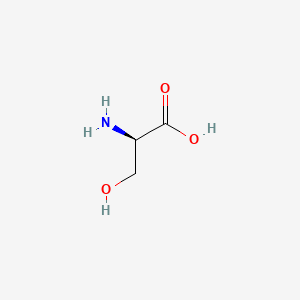 |
0.185 | ||
| ENC000600 | 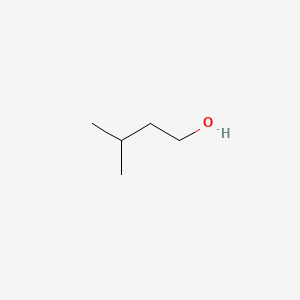 |
0.364 | D0K4MH | 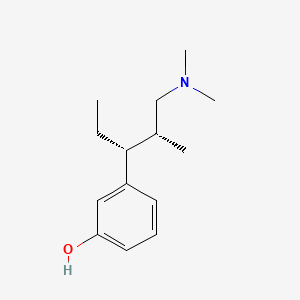 |
0.184 | ||
| ENC000416 | 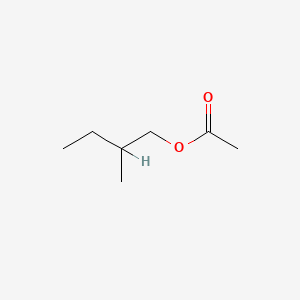 |
0.357 | D0A4JK | 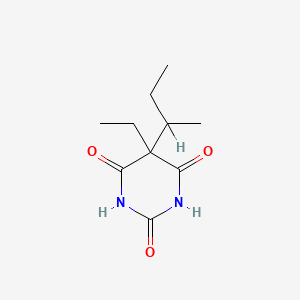 |
0.174 | ||