NPs Basic Information
|
Name |
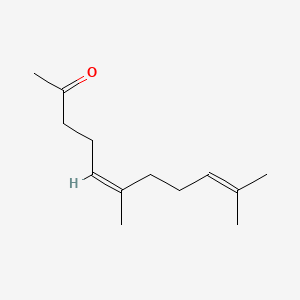
Nerylacetone
|
| Molecular Formula | C13H22O | |
| IUPAC Name* |
(5Z)-6,10-dimethylundeca-5,9-dien-2-one
|
|
| SMILES |
CC(=CCC/C(=C\CCC(=O)C)/C)C
|
|
| InChI |
InChI=1S/C13H22O/c1-11(2)7-5-8-12(3)9-6-10-13(4)14/h7,9H,5-6,8,10H2,1-4H3/b12-9-
|
|
| InChIKey |
HNZUNIKWNYHEJJ-XFXZXTDPSA-N
|
|
| Synonyms |
Nerylacetone; cis-Geranylacetone; 3879-26-3; (5Z)-6,10-dimethylundeca-5,9-dien-2-one; (Z)-Geranylacetone; (E)-geranyl acetone; (Z)-6,10-dimethylundeca-5,9-dien-2-one; KZ9PY9FK2P; 5,9-Undecadien-2-one, 6,10-dimethyl-, (Z)-; 5,9-Undecadien-2-one,6,10-dimethyl-, (5Z)-; 5,9-Undecadien-2-one, 6,10-dimethyl-, cis-; 5,9-Undecadien-2-one, 6,10-dimethyl-,(Z)-; 6,10-Dimethylundecadien-2-one; geranylaceton; Geranyl-Acetone; 6,10-dimethyl-5,9-undecadiene-2-one; 6,10-Dimethylundeca-5,9-dien-2-one; (5E)-6,10-Dimethyl-5,9-undecadien-2-one; UNII-KZ9PY9FK2P; SCHEMBL790270; FEMA 3542; 6,10-Dimethyl-Undecadien-2-one; 68228-05-7; ZINC15121415; AKOS024307561; cis-6,10-dimethyl-5,9-undecadien-2-one; trans-2,6-Dimethyl-2,6-undecadien-2-one; (E)-6,10-dimethyl-5,9- undecadien-2-one; 6,10-Dimethyl-(5E)-5,9-Undecadien-2-one; 6,10-dimethyl-5,9-undecadien-2-one, (E); (5Z)-6,10-dimethyl-undeca-5,9-dien-2-one; F73615; (5Z)-6,10-Dimethyl-5,9-undecadien-2-one #; Q67879718
|
|
| CAS | 3879-26-3 | |
| PubChem CID | 1713001 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 194.31 | ALogp: | 3.7 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 0 |
| Heavy Atoms: | 14 | QED Weighted: | 0.562 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.589 | MDCK Permeability: | 0.00002040 |
| Pgp-inhibitor: | 0.802 | Pgp-substrate: | 0.016 |
| Human Intestinal Absorption (HIA): | 0.015 | 20% Bioavailability (F20%): | 0.992 |
| 30% Bioavailability (F30%): | 0.831 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.959 | Plasma Protein Binding (PPB): | 96.94% |
| Volume Distribution (VD): | 2.34 | Fu: | 3.17% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.669 | CYP1A2-substrate: | 0.735 |
| CYP2C19-inhibitor: | 0.297 | CYP2C19-substrate: | 0.861 |
| CYP2C9-inhibitor: | 0.121 | CYP2C9-substrate: | 0.938 |
| CYP2D6-inhibitor: | 0.046 | CYP2D6-substrate: | 0.851 |
| CYP3A4-inhibitor: | 0.054 | CYP3A4-substrate: | 0.198 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.109 | Half-life (T1/2): | 0.845 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.022 | Human Hepatotoxicity (H-HT): | 0.884 |
| Drug-inuced Liver Injury (DILI): | 0.034 | AMES Toxicity: | 0.002 |
| Rat Oral Acute Toxicity: | 0.009 | Maximum Recommended Daily Dose: | 0.03 |
| Skin Sensitization: | 0.862 | Carcinogencity: | 0.326 |
| Eye Corrosion: | 0.868 | Eye Irritation: | 0.976 |
| Respiratory Toxicity: | 0.016 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001466 |  |
0.725 | D09XWD |  |
0.569 | ||
| ENC001465 |  |
0.725 | D05XQE | 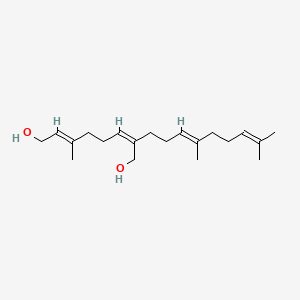 |
0.492 | ||
| ENC001717 | 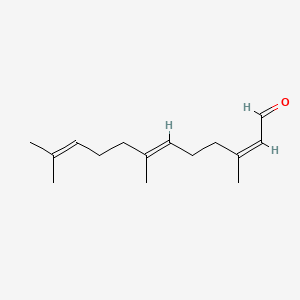 |
0.702 | D03VFL | 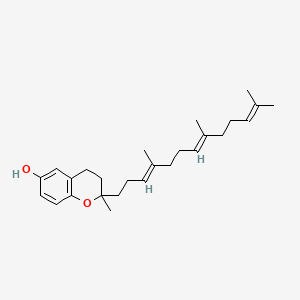 |
0.386 | ||
| ENC002413 | 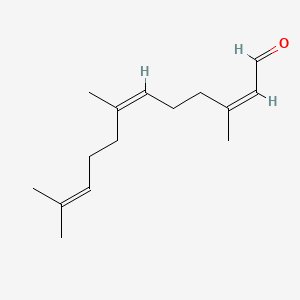 |
0.702 | D0M1PQ |  |
0.313 | ||
| ENC001096 |  |
0.667 | D0Q6DX |  |
0.206 | ||
| ENC001462 |  |
0.667 | D06BLQ | 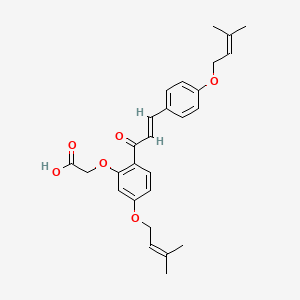 |
0.206 | ||
| ENC001464 | 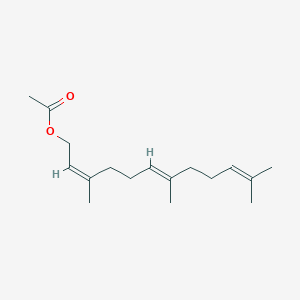 |
0.630 | D0X7XG | 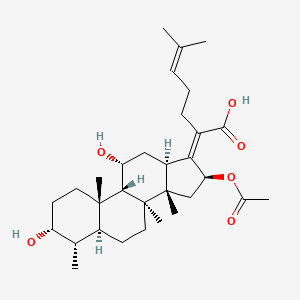 |
0.193 | ||
| ENC000314 | 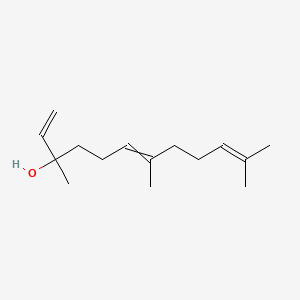 |
0.549 | D0Q9HF | 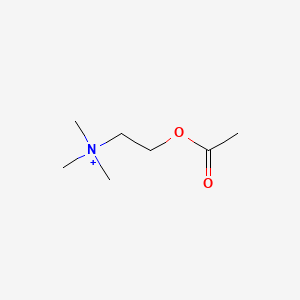 |
0.192 | ||
| ENC001606 | 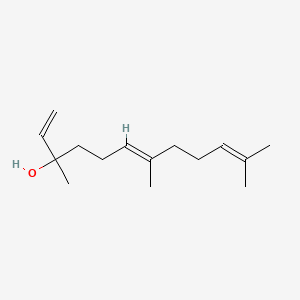 |
0.549 | D0OL6O | 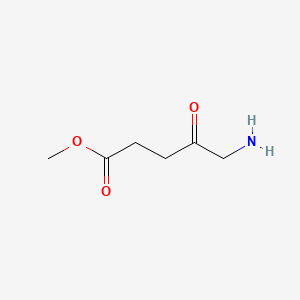 |
0.189 | ||
| ENC001719 | 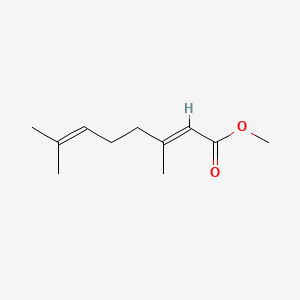 |
0.543 | D0Q7ZQ | 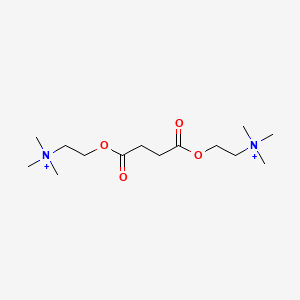 |
0.187 | ||