NPs Basic Information
|
Name |
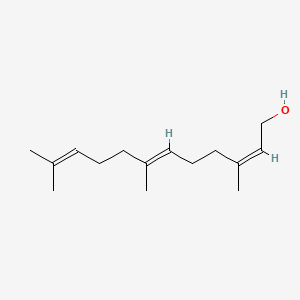
(2Z,6E)-Farnesol
|
| Molecular Formula | C15H26O | |
| IUPAC Name* |
(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol
|
|
| SMILES |
CC(=CCC/C(=C/CC/C(=C\CO)/C)/C)C
|
|
| InChI |
InChI=1S/C15H26O/c1-13(2)7-5-8-14(3)9-6-10-15(4)11-12-16/h7,9,11,16H,5-6,8,10,12H2,1-4H3/b14-9+,15-11-
|
|
| InChIKey |
CRDAMVZIKSXKFV-PVMFERMNSA-N
|
|
| Synonyms |
(Z,E)-Farnesol; (2Z,6E)-Farnesol; cis,trans-Farnesol; 2-cis,6-trans-Farnesol; 3790-71-4; (2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol; (2-cis,6-trans)-farnesol; (Z,E)-3,7,11-Trimethyl-2,6,10-dodecatrien-1-ol; Farnesol, (2Z,6E)-; SQ4TI19PXT; (2Z,6E)-3,7,11-trimethyl-2,6,10-dodecatrien-1-ol; 2,6,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, (Z,E)-; (2-cis,6-trans)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol; 3,7,11-trimethyldodeca-2Z,6E,10-trien-1-ol; Farnesyl alcohol; CIS-TRANS-FARNESOL; UNII-SQ4TI19PXT; NSC60597; (Z)-Farnesol; cis,trans-.alpha.-Farnesol; SCHEMBL806894; CIS-2-TRANS-6-FARNESOL; CHEBI:16774; DTXSID30274196; 4602-84-0; ZINC13507234; LMPR0103010013; FEMA NO. 2478, (2Z,6E)-; 2-cis,6-trans-Farnesol, >=95.0% (GC); C03220; EN300-7460532; 3,7,11-trimethyl-dodeca-2cis,6trans,10-trien-1-ol; Q27102070; 2,6,10-Dodecatrien-1-ol, 3,7,11-trimethyl-, (2Z,6E)-; N1S
|
|
| CAS | 4602-84-0 | |
| PubChem CID | 1549108 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 222.37 | ALogp: | 4.8 |
| HBD: | 1 | HBA: | 1 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 20.2 | Aromatic Rings: | 0 |
| Heavy Atoms: | 16 | QED Weighted: | 0.606 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.496 | MDCK Permeability: | 0.00002280 |
| Pgp-inhibitor: | 0.389 | Pgp-substrate: | 0.01 |
| Human Intestinal Absorption (HIA): | 0.024 | 20% Bioavailability (F20%): | 0.979 |
| 30% Bioavailability (F30%): | 0.97 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.977 | Plasma Protein Binding (PPB): | 97.74% |
| Volume Distribution (VD): | 3.191 | Fu: | 1.97% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.601 | CYP1A2-substrate: | 0.415 |
| CYP2C19-inhibitor: | 0.198 | CYP2C19-substrate: | 0.345 |
| CYP2C9-inhibitor: | 0.149 | CYP2C9-substrate: | 0.904 |
| CYP2D6-inhibitor: | 0.215 | CYP2D6-substrate: | 0.322 |
| CYP3A4-inhibitor: | 0.093 | CYP3A4-substrate: | 0.201 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.195 | Half-life (T1/2): | 0.87 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.012 | Human Hepatotoxicity (H-HT): | 0.733 |
| Drug-inuced Liver Injury (DILI): | 0.011 | AMES Toxicity: | 0.001 |
| Rat Oral Acute Toxicity: | 0.003 | Maximum Recommended Daily Dose: | 0.081 |
| Skin Sensitization: | 0.955 | Carcinogencity: | 0.046 |
| Eye Corrosion: | 0.391 | Eye Irritation: | 0.968 |
| Respiratory Toxicity: | 0.008 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001096 | 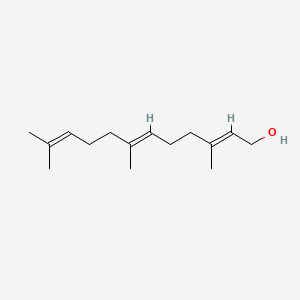 |
1.000 | D05XQE |  |
0.717 | ||
| ENC001717 |  |
0.686 | D09XWD | 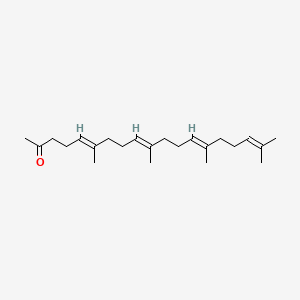 |
0.543 | ||
| ENC001464 | 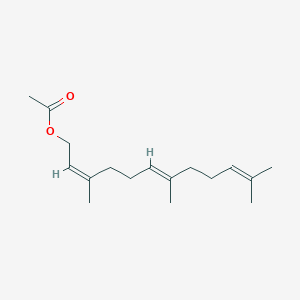 |
0.679 | D03VFL | 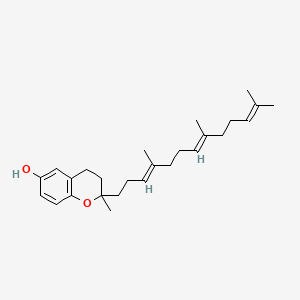 |
0.476 | ||
| ENC001465 | 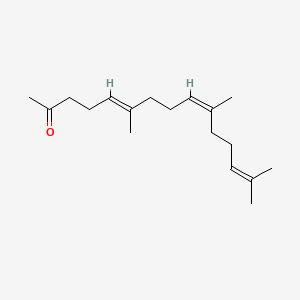 |
0.679 | D0M1PQ | 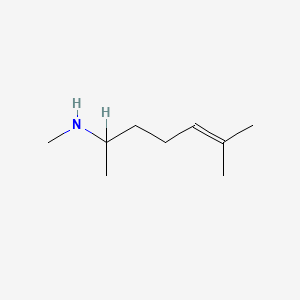 |
0.278 | ||
| ENC001467 | 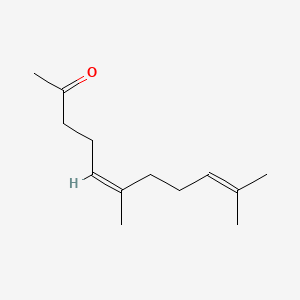 |
0.667 | D01ZUA | 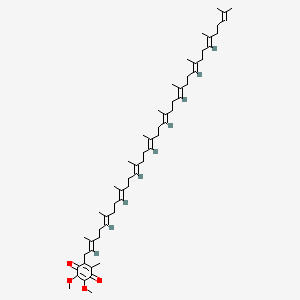 |
0.213 | ||
| ENC001716 | 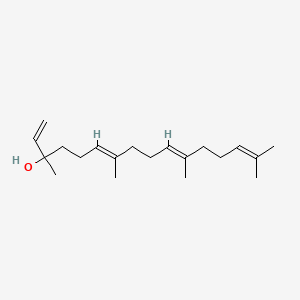 |
0.650 | D0S7WX | 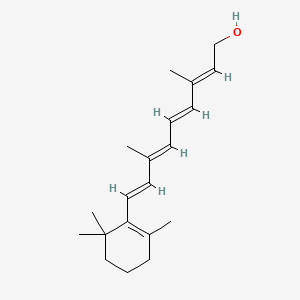 |
0.205 | ||
| ENC000314 | 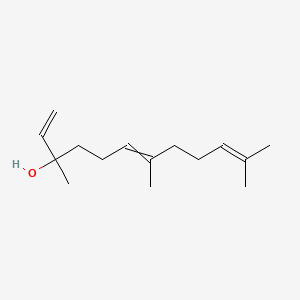 |
0.545 | D06BLQ | 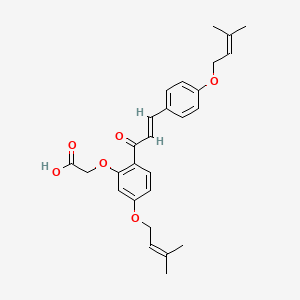 |
0.195 | ||
| ENC001664 | 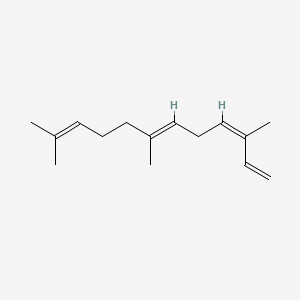 |
0.537 | D0X7XG | 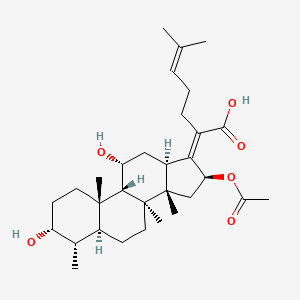 |
0.174 | ||
| ENC006119 | 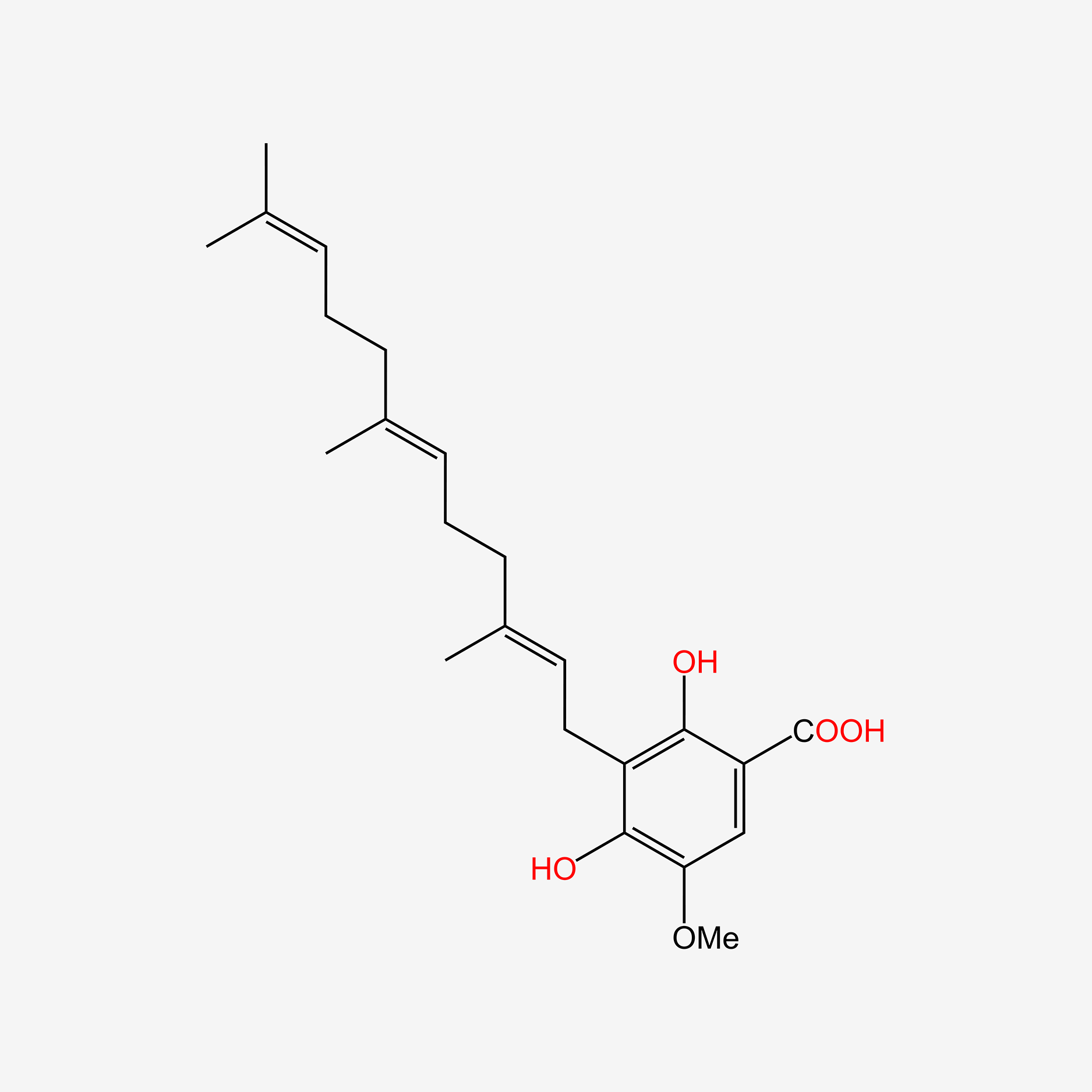 |
0.494 | D0UE9X | 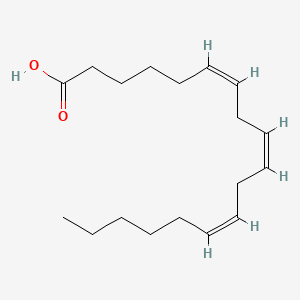 |
0.163 | ||
| ENC001649 | 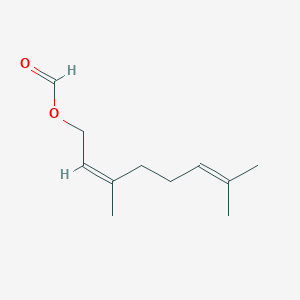 |
0.472 | D04FBR |  |
0.161 | ||